
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,528,884 – 9,529,109 |
| Length | 225 |
| Max. P | 0.999931 |

| Location | 9,528,884 – 9,528,989 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -12.18 |
| Energy contribution | -14.46 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.35 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528884 105 + 22407834 GGUGGAAGUCCUAAGGAUAAGCCUGUAGAAAGUUCU------------GGAGCAAGUCCUGGAGAAAAACCA---GCAGGUAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCU ..((((...(((.(((((..((.(.((((....)))------------).)))..)))))((.......)).---..)))...))))...(((..(((((((.....)))))))...))) ( -29.30) >DroSec_CAF1 36491 108 + 1 GGUGGAAGUCCUAAGGAUAAGCCUGUAGAAAGUUCU------------GGAGGAAAUCCUGGAGCAAAACCAGGAGCAGGUAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCU ..(((((..(((.........(((.((((....)))------------).)))...((((((.......))))))..)))..)))))...(((..(((((((.....)))))))...))) ( -35.50) >DroSim_CAF1 39604 105 + 1 GGUGGAAGUCCUAAGGAUAAGCCUGUAGAAAGUUCU------------GGAGGAAAUCCUGCAGCAAAACCU---GCAGGUAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCU ..(((((((((...))))..((((((((......((------------(.(((....))).)))......))---)))))).)))))...(((..(((((((.....)))))))...))) ( -38.00) >DroEre_CAF1 40389 117 + 1 GCAGGAAGUCCUGAGAGCAAGCCAGCAGAUAGUUCUGGCGGAGCAGCUGGAGGAAGUCCUGCAGGGAAACCU---GUAGUAGGUCCAGAAGGAAAACCAGGAGAUAAUUCUGGCAAAGGC .((((....)))).......(((..........((..((......))..))(((..(.(((((((....)))---)))).)..)))..........((((((.....))))))....))) ( -41.70) >DroYak_CAF1 44576 87 + 1 GGGGGAAGUCCUCAGGUUAAGCCUGUAGAAGGUUCU------------GAAAGAAAUCCUGGGGCCAAACCA---GCAGGACGUCCUGAGAGCAAACCAGGA------------------ ..((...(..((((((.....(((((.(..((((((------------(..(....)..)))))))....).---)))))....))))))..)...))....------------------ ( -27.80) >consensus GGUGGAAGUCCUAAGGAUAAGCCUGUAGAAAGUUCU____________GGAGGAAAUCCUGGAGCAAAACCA___GCAGGUAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCU ...(((.((((...))))..(((((((((....)))............(((.....)))................))))))..))).........(((((((.....)))))))...... (-12.18 = -14.46 + 2.28)



| Location | 9,528,884 – 9,528,989 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -9.75 |
| Energy contribution | -11.87 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.33 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528884 105 - 22407834 AGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGACUACCUGC---UGGUUUUUCUCCAGGACUUGCUCC------------AGAACUUUCUACAGGCUUAUCCUUAGGACUUCCACC (((...(((((((.....)))))))..))).((((((..(((...---.))).....)))((((...(((..------------(((....)))...)))...))))..)))........ ( -25.40) >DroSec_CAF1 36491 108 - 1 AGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUACCUGCUCCUGGUUUUGCUCCAGGAUUUCCUCC------------AGAACUUUCUACAGGCUUAUCCUUAGGACUUCCACC (((...(((((((.....)))))))..)))....((((..((((.((((((.......))))))...(((..------------(((....)))..)))........))))..))))... ( -29.90) >DroSim_CAF1 39604 105 - 1 AGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUACCUGC---AGGUUUUGCUGCAGGAUUUCCUCC------------AGAACUUUCUACAGGCUUAUCCUUAGGACUUCCACC (((...(((((((.....)))))))..)))....((((..(((((---((......)))))))..))))...------------.(((..(((((.(((.....)))))))).))).... ( -31.80) >DroEre_CAF1 40389 117 - 1 GCCUUUGCCAGAAUUAUCUCCUGGUUUUCCUUCUGGACCUACUAC---AGGUUUCCCUGCAGGACUUCCUCCAGCUGCUCCGCCAGAACUAUCUGCUGGCUUGCUCUCAGGACUUCCUGC ((((..(((((.........)))))..(((....)))........---))))......((((((..((((..(((....(((.((((....)))).)))...)))...))))..)))))) ( -33.60) >DroYak_CAF1 44576 87 - 1 ------------------UCCUGGUUUGCUCUCAGGACGUCCUGC---UGGUUUGGCCCCAGGAUUUCUUUC------------AGAACCUUCUACAGGCUUAACCUGAGGACUUCCCCC ------------------((((((.......)))))).(((((.(---.((((.((((...(((.(((....------------.)))...)))...)))).)))).))))))....... ( -26.80) >consensus AGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGACUACCUGC___UGGUUUUGCUCCAGGAUUUCCUCC____________AGAACUUUCUACAGGCUUAUCCUUAGGACUUCCACC ......(((((((.....))))))).........(((...((((.....((((((......(((.....)))............)))))).....))))((((....))))...)))... ( -9.75 = -11.87 + 2.12)


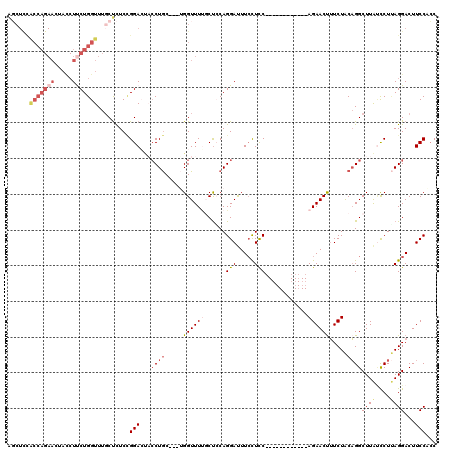
| Location | 9,528,920 – 9,529,029 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -16.59 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528920 109 + 22407834 --------GGAGCAAGUCCUGGAGAAAAACCA---GCAGGUAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGUUGGAGCUGCUGGAGAAAAGCCUGUUGAUAUUC --------(((.....)))...........((---((((((..(((((..(((...((((..((((((((.....))))))))....)))).))).))))).....))))))))...... ( -36.60) >DroSec_CAF1 36527 112 + 1 --------GGAGGAAAUCCUGGAGCAAAACCAGGAGCAGGUAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUAAAGGUGGAGCUACUGGACAAAAUCCUAUUGAAAUUC --------..((((..((((((.......))))))........(((((..(((..(((....((((((((.....))))))))...)))...))).))))).....)))).......... ( -35.50) >DroSim_CAF1 39640 109 + 1 --------GGAGGAAAUCCUGCAGCAAAACCU---GCAGGUAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGGUGGAGCUACUGGAGAAAAUCCUGUUGAAAUUC --------..((((...(((((((......))---)))))..((((((..(((..(((....((((((((.....))))))))...)))...))).))))))....)))).......... ( -39.70) >DroEre_CAF1 40429 114 + 1 GAGCAGCUGGAGGAAGUCCUGCAGGGAAACCU---GUAGUAGGUCCAGAAGGAAAACCAGGAGAUAAUUCUGGCAAAGGCACUGG---GGGAAGUCCCCAGGAUAAGCCUGUAGAAAGUC ..((.((..(((((..(.(((((((....)))---)))).)..)))....((....))..........))..))..((((.((((---((.....)))))).....))))))........ ( -43.30) >consensus ________GGAGGAAAUCCUGCAGCAAAACCA___GCAGGUAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGGUGGAGCUACUGGAGAAAAGCCUGUUGAAAUUC ........(((.....(((.(.(((...(((....((((.(...((((((.....(((....)))..))))))...).))))....)))...))).).))).....)))........... (-16.59 = -19.40 + 2.81)


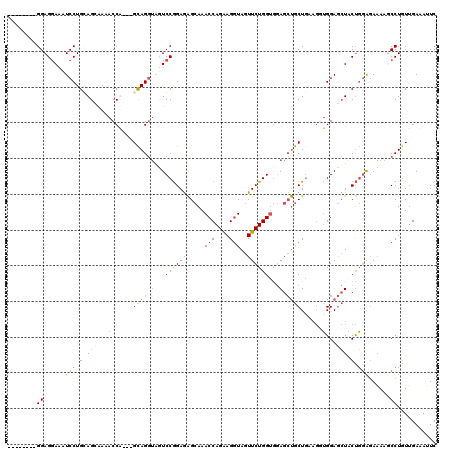
| Location | 9,528,920 – 9,529,029 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528920 109 - 22407834 GAAUAUCAACAGGCUUUUCUCCAGCAGCUCCAACUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGACUACCUGC---UGGUUUUUCUCCAGGACUUGCUCC-------- ...........((.......(((((((.(((......(((((....(((((((.....))))))))))))....)))....))))---))).......)).(((.....)))-------- ( -27.34) >DroSec_CAF1 36527 112 - 1 GAAUUUCAAUAGGAUUUUGUCCAGUAGCUCCACCUUUAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUACCUGCUCCUGGUUUUGCUCCAGGAUUUCCUCC-------- ..........((((....((((.(.(((...(((...((((((...(((((((.....)))))))..)).....((....))))))...)))...))).).)))).))))..-------- ( -30.00) >DroSim_CAF1 39640 109 - 1 GAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUACCUGC---AGGUUUUGCUGCAGGAUUUCCUCC-------- ...........(((............(((........)))(((...(((((((.....)))))))..))).)))((((..(((((---((......)))))))..))))...-------- ( -33.00) >DroEre_CAF1 40429 114 - 1 GACUUUCUACAGGCUUAUCCUGGGGACUUCCC---CCAGUGCCUUUGCCAGAAUUAUCUCCUGGUUUUCCUUCUGGACCUACUAC---AGGUUUCCCUGCAGGACUUCCUCCAGCUGCUC ..........((((.....((((((.....))---)))).))))..(((((.........))))).......(((((.......(---(((....))))..((....)))))))...... ( -31.20) >consensus GAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUACCUGC___AGGUUUUGCUCCAGGACUUCCUCC________ ......................(((((.(((.....((((((..(((((((((.....)))))))........(((.....))).....))..))))))..))).))))).......... (-15.35 = -15.97 + 0.62)

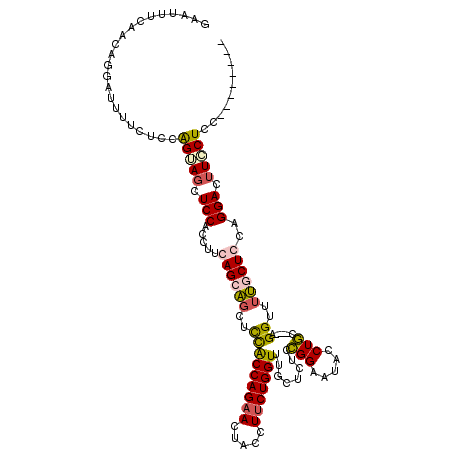
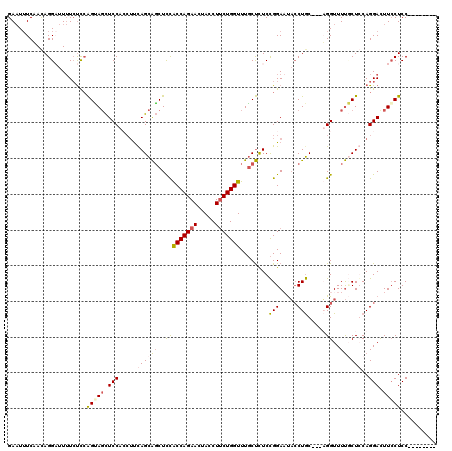
| Location | 9,528,949 – 9,529,069 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528949 120 + 22407834 UAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGUUGGAGCUGCUGGAGAAAAGCCUGUUGAUAUUCCACAACAAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGG ...(((((..(((...((((..((((((((.....))))))))....)))).))).)))))....(((((((((........)))).....((((((.....))))))...))))).... ( -39.60) >DroSec_CAF1 36559 120 + 1 UAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUAAAGGUGGAGCUACUGGACAAAAUCCUAUUGAAAUUCCACCAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCGGG ....((.(((..(...(((((((((..(((((((((((((.(.......).)))....(((.....))).........))))))))))..))).........))))))..)...))).)) ( -39.10) >DroSim_CAF1 39669 120 + 1 UAUUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGGUGGAGCUACUGGAGAAAAUCCUGUUGAAAUUCCACCAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGG ....((((((..(...(((((((((..(((((((((((((.(.......).))).((.(((.....))).))......))))))))))..))).........))))))..)...)))))) ( -42.60) >DroEre_CAF1 40466 117 + 1 AGGUCCAGAAGGAAAACCAGGAGAUAAUUCUGGCAAAGGCACUGG---GGGAAGUCCCCAGGAUAAGCCUGUAGAAAGUCCUGAGAGCAAGCCAGGAGGUAGUUCUGGCGGAGCAGUUGG .(.(((...((((...((((((.....))))))...((((.((((---((.....)))))).....))))........))))........(((((((.....)))))))))).)...... ( -43.20) >consensus UAGUCCGGAGAGCAAACCAGAAGGUAGUUCUGGUGGAGCUGCUGAAGGUGGAGCUACUGGAGAAAAGCCUGUUGAAAUUCCACAAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGG ....((((((......((((((.((..(((((((..((((.(.......).))))...........((((..((......))..))))..))))))).))..))))))......)))))) (-27.55 = -27.80 + 0.25)

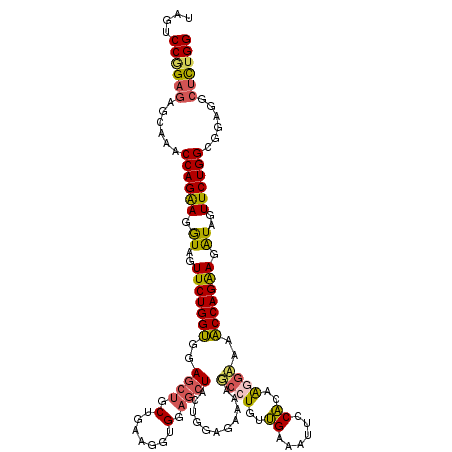
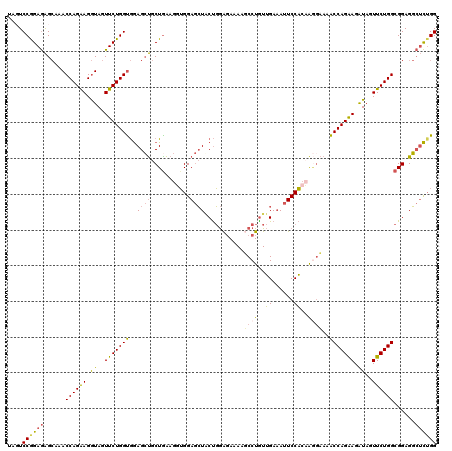
| Location | 9,528,949 – 9,529,069 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528949 120 - 22407834 CCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUUGUUGUGGAAUAUCAACAGGCUUUUCUCCAGCAGCUCCAACUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGACUA ..((((((...(((((((.....)))))))...(((((........)))))))))))..(((....(((........)))(((...(((((((.....)))))))..)))....)))... ( -34.70) >DroSec_CAF1 36559 120 - 1 CCCGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAAUAGGAUUUUGUCCAGUAGCUCCACCUUUAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUA ((.((((..(((((((((.....)))))))....((((((((.........((((...)))).((.(((........))).))))))))))...........))...))))...)).... ( -36.40) >DroSim_CAF1 39669 120 - 1 CCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUA (((((((..(((((((((.....)))))))....((((((((.........(((.....))).((.(((........))).))))))))))...........))...)))))..)).... ( -37.50) >DroEre_CAF1 40466 117 - 1 CCAACUGCUCCGCCAGAACUACCUCCUGGCUUGCUCUCAGGACUUUCUACAGGCUUAUCCUGGGGACUUCCC---CCAGUGCCUUUGCCAGAAUUAUCUCCUGGUUUUCCUUCUGGACCU (((...((...(((((.........)))))..))....((((........((((.....((((((.....))---)))).))))..(((((.........)))))..))))..))).... ( -34.90) >consensus CCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGCAGCUCCACCAGAACUACCUUCUGGUUUGCUCUCCGGAAUA (((((((....(((((((.....(((((((..(((((.(((....))).)))))........(((.(((........))).)))..))))))).....)))))))..)))))..)).... (-26.95 = -26.70 + -0.25)

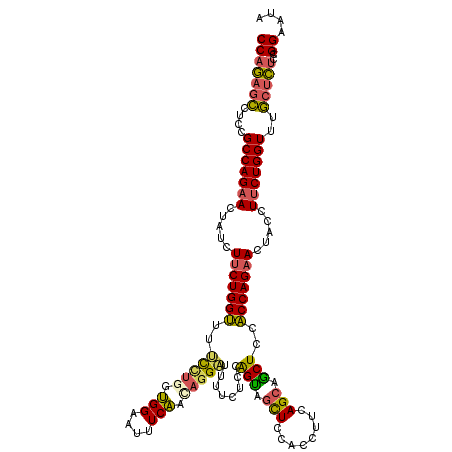
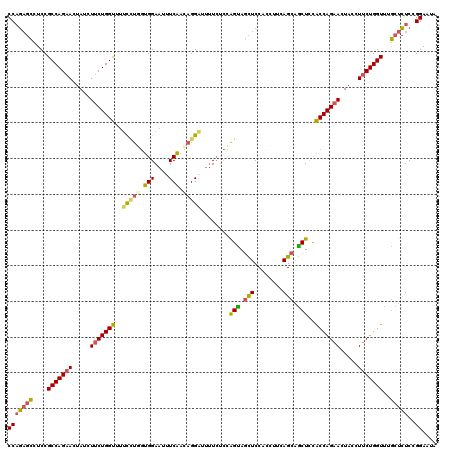
| Location | 9,528,989 – 9,529,109 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -20.71 |
| Energy contribution | -22.15 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528989 120 + 22407834 GCUGAAGUUGGAGCUGCUGGAGAAAAGCCUGUUGAUAUUCCACAACAAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGGGGGAAGUCCUCAGAAUAAGCCUGUAGAAAGUUCUAAAGGA (((...((..((((((((((((......))((((........)))).....))))....))))))..))...)))(((((((.....))))))).....(((.((((....)))).))). ( -37.40) >DroSec_CAF1 36599 119 + 1 GCUAAAGGUGGAGCUACUGGACAAAAUCCUAUUGAAAUUCCACCAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCGGGGGGAAGUCCCAAGGAUAAGCCUGUAGAAAGUCCUAAAGG- .((..(((.....((((.((.....(((((.((((..((((.((((((....(....)....)))))).))))..)))).(((....))).)))))...)).)))).....)))..)).- ( -35.20) >DroSim_CAF1 39709 119 + 1 GCUGAAGGUGGAGCUACUGGAGAAAAUCCUGUUGAAAUUCCACCAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGGGGGAAGUCCUCAGAAUAAGCCUGUAGAAAGUCCUAAAGG- ......(((((((..((.(((.....))).)).....)))))))((((...((((((.....)))))).(.(((((((((((.....)))))))....)))).)......)))).....- ( -39.60) >DroEre_CAF1 40506 115 + 1 ACUGG---GGGAAGUCCCCAGGAUAAGCCUGUAGAAAGUCCUGAGAGCAAGCCAGGAGGUAGUUCUGGCGGAGCAGUUGGAGGAAGUCCUGGAGGGAGACCUGUAGUAAAACCAGAAG-- ((((.---(((...((((((((((...(((.(((....(((((....)).(((((((.....))))))))))....))).)))..))))))).)))...))).))))...........-- ( -42.50) >consensus GCUGAAGGUGGAGCUACUGGAGAAAAGCCUGUUGAAAUUCCACAAGGAAAACCAGAAGAUAGUUCUGGCGGAGGCUCUGGGGGAAGUCCUCAGAAUAAGCCUGUAGAAAGUCCUAAAGG_ .(((.((((((((((.(((.......((((..((......))..))))...((((((.....))))))))).)))))).((((....)))).......)))).))).............. (-20.71 = -22.15 + 1.44)


| Location | 9,528,989 – 9,529,109 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9528989 120 - 22407834 UCCUUUAGAACUUUCUACAGGCUUAUUCUGAGGACUUCCCCCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUUGUUGUGGAAUAUCAACAGGCUUUUCUCCAGCAGCUCCAACUUCAGC .(((.((((....)))).)))......((((((....))....((((....(((((((.....)))))))...(((((..(((..((....))...)))..))))))))).....)))). ( -28.70) >DroSec_CAF1 36599 119 - 1 -CCUUUAGGACUUUCUACAGGCUUAUCCUUGGGACUUCCCCCCGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAAUAGGAUUUUGUCCAGUAGCUCCACCUUUAGC -.((..(((.....((((.(((..(((((.(((....)))...(((..(((((((((((((.....)))))...))))))))..)))...)))))...)))..)))).....)))..)). ( -35.50) >DroSim_CAF1 39709 119 - 1 -CCUUUAGGACUUUCUACAGGCUUAUUCUGAGGACUUCCCCCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGC -.((..(((.....((((.(((....((((.((......)))))))))(((((((((((((.....)))))...)))))))).........(((.....))).)))).....)))..)). ( -32.80) >DroEre_CAF1 40506 115 - 1 --CUUCUGGUUUUACUACAGGUCUCCCUCCAGGACUUCCUCCAACUGCUCCGCCAGAACUACCUCCUGGCUUGCUCUCAGGACUUUCUACAGGCUUAUCCUGGGGACUUCCC---CCAGU --...((((........((((....(((..((((..((((......((...(((((.........)))))..))....))))..))))..))).....))))(((....)))---)))). ( -32.90) >consensus _CCUUUAGGACUUUCUACAGGCUUAUCCUGAGGACUUCCCCCAGAGCCUCCGCCAGAACUAUCUUCUGGUUUUCCUGGUGGAAUUUCAACAGGAUUUUCUCCAGUAGCUCCACCUUCAGC ...................((......(((((((.................(((((((.....)))))))..(((((.(((....))).)))))...))))))).....))......... (-17.01 = -17.20 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:58 2006