
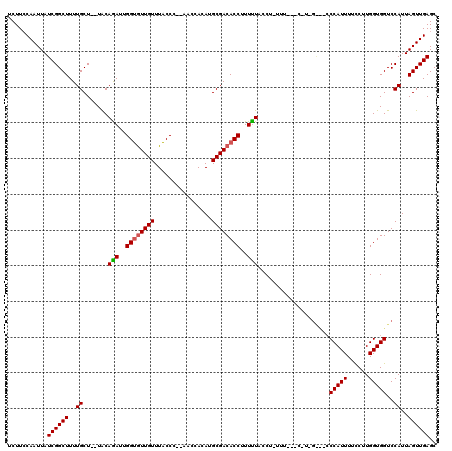
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,483,556 – 9,483,704 |
| Length | 148 |
| Max. P | 0.837653 |

| Location | 9,483,556 – 9,483,664 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
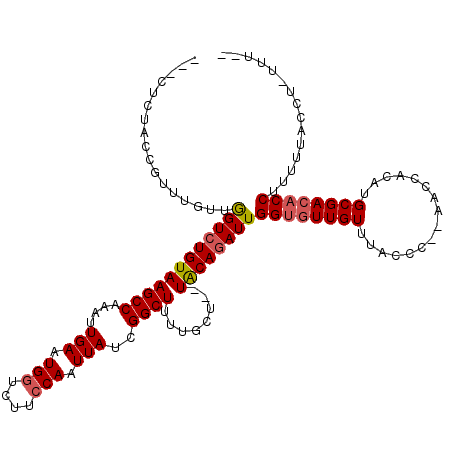
>2L_DroMel_CAF1 9483556 108 - 22407834 UCUUGCAAUUAUCGGCUUUUGCU--UACAAAUUGGUAUUGUUUACCC--AACCACAUGCGACACCUUUUUGCC--------CAUAGGUAGCCAUUUUCCUUGGUGGUCCAUUAGUUGAGC .....((((((..((....((((--((((((..(((.((((......--........)))).)))..))))..--------..))))))((((((......))))))))..))))))... ( -21.24) >DroEre_CAF1 42050 118 - 1 UCUUCCAAUUAUCGGCUUUUGCUUUUGCAGAUUGGCGUUGUUUGCCC--ACCCAAAUGCGACACCUUUCUACCUCUUUUCCCUUCGACUCCCAUUUUUCUUGGUGGUCCAUUAGUUGAGC ...........((((((..((.......(((..((.((((((((...--...)))..))))).))..)))...............(((..(((.......)))..)))))..)))))).. ( -25.30) >DroYak_CAF1 42678 108 - 1 UCUUCCAAUUAUCGGCUUUUGCU--UACAGAUUGGUGUUGUUUACCCCAAACCACAUGCGACACCUUUUUCCCUUUUUC----------CCCAUUUUCCUUGGUGGUCCAUUAGUUGAGC ...........((((((..((..--...(((..((((((((................))))))))..)))........(----------((((.......))).))..))..)))))).. ( -21.49) >consensus UCUUCCAAUUAUCGGCUUUUGCU__UACAGAUUGGUGUUGUUUACCC__AACCACAUGCGACACCUUUUUACCU_UUU___C_U_G___CCCAUUUUCCUUGGUGGUCCAUUAGUUGAGC ...........((((((..((.......(((..((((((((................))))))))..)))....................(((((......)))))..))..)))))).. (-19.10 = -19.32 + 0.22)


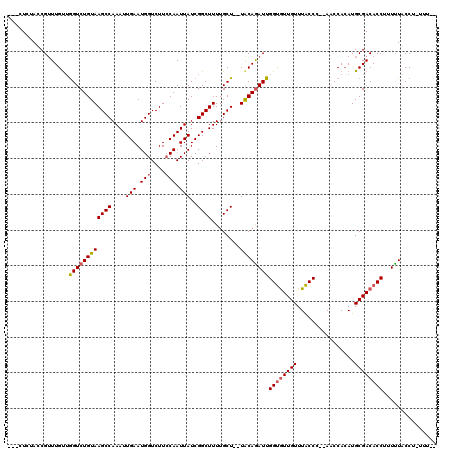
| Location | 9,483,595 – 9,483,704 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9483595 109 - 22407834 UCUCUCUACCGAUUGUUGGUCUGUAAGCCAAAUUGAAUGGUCUUGCAAUUAUCGGCUUUUGCU--UACAAAUUGGUAUUGUUUACCC--AACCACAUGCGACACCUUUUUGCC------- .........(((..((((((.(((((((((.......))).))))))...))))))..)))..--..((((..(((.((((......--........)))).)))..))))..------- ( -22.14) >DroEre_CAF1 42090 115 - 1 ---CUUUACCGUUUGUUAGUCUGUAAGCCAAAUUGAAUGGUCUUCCAAUUAUCGGCUUUUGCUUUUGCAGAUUGGCGUUGUUUGCCC--ACCCAAAUGCGACACCUUUCUACCUCUUUUC ---...........(((((((((((((((....(((.(((....))).)))..))))........)))))))))))((((((((...--...)))..))))).................. ( -23.70) >DroYak_CAF1 42709 113 - 1 ----UCAAGCGUUUGCGGGUCUGUAAGCCAAAUUGAAUGGUCUUCCAAUUAUCGGCUUUUGCU--UACAGAUUGGUGUUGUUUACCCCAAACCACAUGCGACACCUUUUUCCCUUUUUC- ----....((....))((((((((((((.(((((((.(((....)))....)))).))).)))--))))))..((((((((................)))))))).....)))......- ( -30.99) >consensus ___CUCUACCGUUUGUUGGUCUGUAAGCCAAAUUGAAUGGUCUUCCAAUUAUCGGCUUUUGCU__UACAGAUUGGUGUUGUUUACCC__AACCACAUGCGACACCUUUUUACCU_UUU__ .................((((((((((((....(((.(((....))).)))..))))........))))))))((((((((................))))))))............... (-17.63 = -18.52 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:34 2006