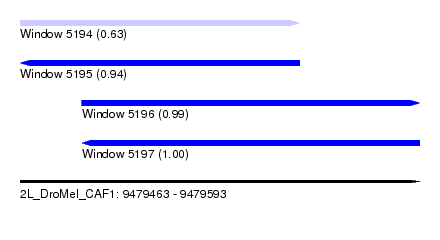
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,479,463 – 9,479,593 |
| Length | 130 |
| Max. P | 0.998402 |

| Location | 9,479,463 – 9,479,554 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 98.34 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -33.92 |
| Energy contribution | -35.93 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9479463 91 + 22407834 AUCAGUGUGUGGGUGUGGAAGGUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGAGCAAAAAAAGAA (((((((((..((..........))..)))))))))((((((((((....))))))))))((((..(((.....))))))).......... ( -36.20) >DroSec_CAF1 40544 90 + 1 AUCAGUGUGUGGGUGUGGAAGGUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGAGCAAA-AAAGAA (((((((((..((..........))..)))))))))((((((((((....))))))))))((((..(((.....)))))))...-...... ( -36.20) >DroEre_CAF1 41372 90 + 1 AUCAGUGUGUGGGUGUGGAAGGUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAGGAGCGGAGCAAA-AAAGAA (((((((((..((..........))..)))))))))((((((((((....))))))))))((((..(((.....)))))))...-...... ( -36.20) >DroYak_CAF1 41887 90 + 1 AUCAGUGUGUGGGUGUGGAAGGUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGUGCGGAGCAAA-AAAGAA (((((((((..((..........))..)))))))))((((((((((....))))))))))((((..(((...)))..))))...-...... ( -35.40) >consensus AUCAGUGUGUGGGUGUGGAAGGUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGAGCAAA_AAAGAA (((((((((..((..........))..)))))))))((((((((((....))))))))))((((..(((.....))))))).......... (-33.92 = -35.93 + 2.00)


| Location | 9,479,463 – 9,479,554 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 98.34 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9479463 91 - 22407834 UUCUUUUUUUGCUCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGACCUUCCACACCCACACACUGAU ....((((.(((..........))).)))).((((((((((....))))))))))..(((((....((..........))....))))).. ( -22.10) >DroSec_CAF1 40544 90 - 1 UUCUUU-UUUGCUCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGACCUUCCACACCCACACACUGAU ...(((-(.(((..........))).)))).((((((((((....))))))))))..(((((....((..........))....))))).. ( -22.00) >DroEre_CAF1 41372 90 - 1 UUCUUU-UUUGCUCCGCUCCUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGACCUUCCACACCCACACACUGAU ......-........(.((((.((((.....((((((((((....))))))))))....))))..)))).).................... ( -22.20) >DroYak_CAF1 41887 90 - 1 UUCUUU-UUUGCUCCGCACUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGACCUUCCACACCCACACACUGAU ...(((-(.(((..........))).)))).((((((((((....))))))))))..(((((....((..........))....))))).. ( -22.00) >consensus UUCUUU_UUUGCUCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGACCUUCCACACCCACACACUGAU ...............((.....)).......((((((((((....))))))))))..(((((....((..........))....))))).. (-21.52 = -21.52 + 0.00)

| Location | 9,479,483 – 9,479,593 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -20.78 |
| Energy contribution | -23.22 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9479483 110 + 22407834 G---GUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGA--GCAAAAAAAGAAGGUGA-ACGCUGAGGUCGAAUUGGGUUAGAUCGCUGAGGA .---.((((.((((..(..((((((((((....))))))))))..)...))))...((((((--((.......((.(((..-..)))....)).......)))...))))).)))) ( -33.54) >DroSec_CAF1 40564 109 + 1 G---GUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGA--GCAAA-AAAGAAGGUGA-ACGCUGAGGUCGGUUUGGGUUAGAUCGUUGAGGA .---.((((((.((((...((((((((((....))))))))))((((..(((.....)))))--))...-......)))).-)).....((((...........))))....)))) ( -33.00) >DroEre_CAF1 41392 98 + 1 G---GUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAGGAGCGGA--GCAAA-AAAGAAGGUGA-ACGG-----------CGGGUUAGAUCGGAAGGGA .---..(((((.((((...((((((((((....))))))))))((((..(((.....)))))--))...-......)))).-...)-----------)))).....((....)).. ( -32.70) >DroYak_CAF1 41907 98 + 1 G---GUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGUGCGGA--GCAAA-AAAGAAGGUGA-ACGG-----------UGGGUUAGAUCCGAAAGGA .---.((((((.((((...((((((((((....))))))))))((((..(((...)))..))--))...-......)))).-))))-----------.))......(((....))) ( -34.30) >DroAna_CAF1 59551 103 + 1 GUGAGCGCUGUGCCUUGCUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAAACCAAAAGGCAAA-AAAGAAGGUAGCAUUC-----------UCGGUUA-UUCGGAACAGA ....(((((.((((((...((((((((((....))))))))))(((((.....)))))...))))))..-......))).)).(((-----------.(((...-.)))))).... ( -31.70) >consensus G___GUCCUGUGCACUGGUUGACGCCAAAGAAAUUUGGCGUCAGUUUUAUGCGAAGAGCGGA__GCAAA_AAAGAAGGUGA_ACGC___________UGGGUUAGAUCGGAAAGGA ....(..((.((((..(((((((((((((....)))))))))))))...)))).))..)......................................................... (-20.78 = -23.22 + 2.44)

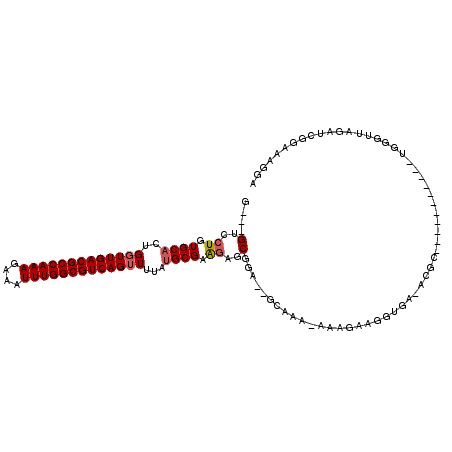

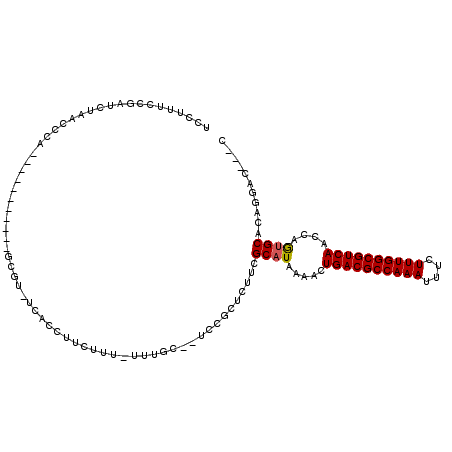
| Location | 9,479,483 – 9,479,593 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9479483 110 - 22407834 UCCUCAGCGAUCUAACCCAAUUCGACCUCAGCGU-UCACCUUCUUUUUUUGC--UCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGAC---C ((((.((((..............(....)((((.-..............)))--).))))....((((.....((((((((((....))))))))))....))))..)))).---. ( -26.16) >DroSec_CAF1 40564 109 - 1 UCCUCAACGAUCUAACCCAAACCGACCUCAGCGU-UCACCUUCUUU-UUUGC--UCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGAC---C ((((...................((.(..((((.-...........-..)))--)..).))...((((.....((((((((((....))))))))))....))))..)))).---. ( -23.94) >DroEre_CAF1 41392 98 - 1 UCCCUUCCGAUCUAACCCG-----------CCGU-UCACCUUCUUU-UUUGC--UCCGCUCCUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGAC---C .....(((..........(-----------(.(.-.((........-..)).--.).)).....((((.....((((((((((....))))))))))....))))...))).---. ( -22.20) >DroYak_CAF1 41907 98 - 1 UCCUUUCGGAUCUAACCCA-----------CCGU-UCACCUUCUUU-UUUGC--UCCGCACUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGAC---C ((((...((.......)).-----------..((-.(((....(((-(.(((--..........))).)))).((((((((((....))))))))))....))).)))))).---. ( -24.30) >DroAna_CAF1 59551 103 - 1 UCUGUUCCGAA-UAACCGA-----------GAAUGCUACCUUCUUU-UUUGCCUUUUGGUUUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAAGCAAGGCACAGCGCUCAC .((((.(((((-.((((((-----------((..((..........-...)).))))))))))).........((((((((((....)))))))))).....)).))))....... ( -30.22) >consensus UCCUUUCCGAUCUAACCCA___________GCGU_UCACCUUCUUU_UUUGC__UCCGCUCUUCGCAUAAAACUGACGCCAAAUUUCUUUGGCGUCAACCAGUGCACAGGAC___C ................................................................((((.....((((((((((....))))))))))....))))........... (-18.26 = -18.30 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:32 2006