| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 987,012 – 987,105 |
| Length | 93 |
| Max. P | 0.556736 |

| Location | 987,012 – 987,105 |
|---|---|
| Length | 93 |
| Sequences | 6 |
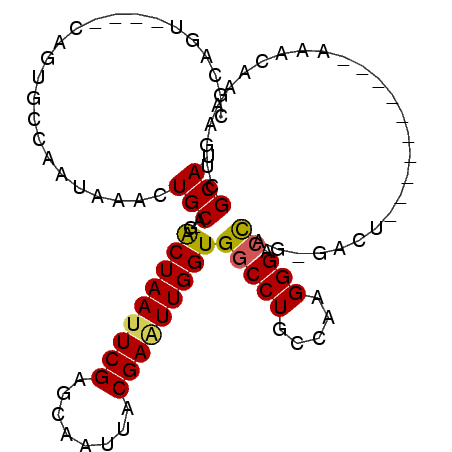
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
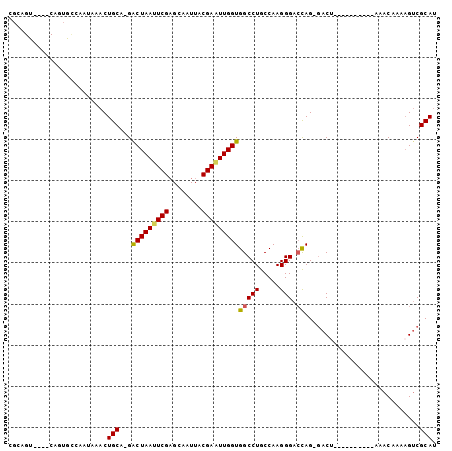
>2L_DroMel_CAF1 987012 93 - 22407834 CGCAGUCAGUCAGUGCCAAUAAACUGCA-GACUAAUUCGAGCAAUUACGAGUUGGUGGCCUGCCAAGGGACCAGGGACU----------AAACAAAAGUCGCAU .(((((..((........))..))))).-.(((((((((........)))))))))(((((.....))).)).(.((((----------.......)))).).. ( -24.70) >DroPse_CAF1 113033 99 - 1 AUCGGA----CACUUUCCAUAAACUGCACGACUAAUUCGUGCAAUUACGAAUUGGUGGCCUGCCAAGGGAAUAG-AACCCCGAAAACUGAAACAAAAGUCGCAU .((((.----..((((((......(((((((.....)))))))........(((((.....))))).)))).))-....))))....((..((....))..)). ( -23.10) >DroSec_CAF1 107923 88 - 1 CGCAGU----CAGUGCCAAUAAACUGCA-GACUAAUUCGAGCAAUUACGAAUUGGUGGCCUGCCAAGGGACCAG-GACU----------AAACAAAAGUCGCAU .(((((----..((....))..))))).-.(((((((((........)))))))))(((((.....))).))..-((((----------.......)))).... ( -24.00) >DroSim_CAF1 108535 88 - 1 CGCAGU----CAGUGCCAAUAAACUGCA-GACUAAUUCGAGCAAUUACGAAUUGGUGGCCUGCCAAGGGACCAG-GACU----------AAACAAAAGUCGCAU .(((((----..((....))..))))).-.(((((((((........)))))))))(((((.....))).))..-((((----------.......)))).... ( -24.00) >DroEre_CAF1 114019 77 - 1 C---------------CAAUAGACUGCC-GGCUAAUUCGAGCCAUUACGAUUUGGUGGCCUGCCAAGGGACCAG-GACU----------AAACAAAUGUCGCAU .---------------.....(((....-((((......))))......(((((.(((((((((...))..)))-).))----------)..)))))))).... ( -17.20) >DroYak_CAF1 114426 78 - 1 U--------------UCAAUAGACUGCA-GACUAAUUCGAGCAAUUACGAAUUGGUGGCCUGCCAAGGGACCAG-GACU----------AAACAAAAGGCGCAC .--------------.........(((.-.(((((((((........))))))))).((((.....((......-..))----------.......))))))). ( -17.50) >consensus CGCAGU____CAGUGCCAAUAAACUGCA_GACUAAUUCGAGCAAUUACGAAUUGGUGGCCUGCCAAGGGACCAG_GACU__________AAACAAAAGUCGCAU ........................(((...(((((((((........)))))))))(((((.....))).))............................))). (-15.80 = -15.75 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:05 2006