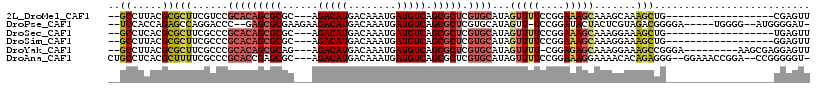
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
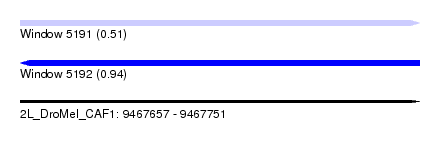
| Location | 9,467,657 – 9,467,751 |
| Length | 94 |
| Max. P | 0.942151 |

| Location | 9,467,657 – 9,467,751 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -15.72 |
| Energy contribution | -17.08 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
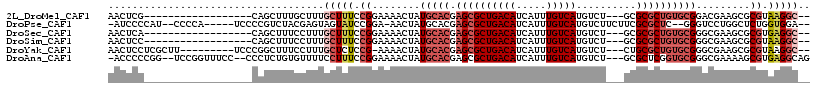
>2L_DroMel_CAF1 9467657 94 + 22407834 AACUCG------------------CAGCUUUGCUUUGCUUUCCGGAAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU---GCGCGCUGUGCGGACGAAGCGCGUAAGGC-- ......------------------..(((((((..(((((((((.....)...(((((.((((.(((((........))))).---))))..))))))))..))))).)))))))-- ( -33.60) >DroPse_CAF1 31709 104 + 1 -AUCCCCAU--CCCCA-----UCCCCGUCUACGAGUAGUAUCCGGA-AACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCUUCUUCGCGCUC--GGGUCCUGGCUCUGGUGGA-- -.((((((.--..(((-----.(((((..(((.....)))..))).-.........((((((((((((.....)))))..........))))))--)))...)))...))).)))-- ( -33.30) >DroSec_CAF1 32411 94 + 1 AACUCA------------------CAGCUUUCCUUUGCUUUCCGGAAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU---GCGCGCUGUGCGGGCGAAGCGCGUGAGGC-- ..((((------------------(((.(((((..........))))).))........(((((((((.....)))).(((((---((((...))))))))).))))))))))..-- ( -36.00) >DroSim_CAF1 32263 94 + 1 AACUCC------------------CAGCUUUCCUUUGCUUUCCGGAAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU---GCGCGCUGUGCGGGCGAAGCGCGUAAGGC-- ..(.((------------------(((.(((((..........))))).))..(((((.((((.(((((........))))).---))))..)))))))).)..((.(....)))-- ( -31.70) >DroYak_CAF1 33120 102 + 1 AACUCCUCGCUU---------UCCCGGCUUUCCUUUGCUCUCCG-AAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU---CUGCGCUGUGCGGGCGAAGCGCGUAAGGC-- ....((((((((---------(((((((........))).....-........((((.(((((.(((((........))))).---..)))))))))))).))))))....))).-- ( -35.40) >DroAna_CAF1 31103 109 + 1 -ACCCCCGG--UCCGGUUUCC--CCCUCUGUGUUUUCCUUUCCGGAAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU---GCGCUCGGUGCGGGCGAAAAGCGUGAGGCAG -....((..--((((.((((.--(((.....(((((((.....)))))))...((((((((((.(((((........))))).---)))))).))))))).))))..)).))))... ( -41.80) >consensus AACUCC__________________CAGCUUUCCUUUGCUUUCCGGAAAACUAUGCACGAGCGCUGACAUCAUUUGUCAUGUCU___GCGCGCUGUGCGGGCGAAGCGCGUAAGGC__ ....................................(((((.((........(((((.((((((((((.....)))))..........)))))))))).........)).))))).. (-15.72 = -17.08 + 1.36)
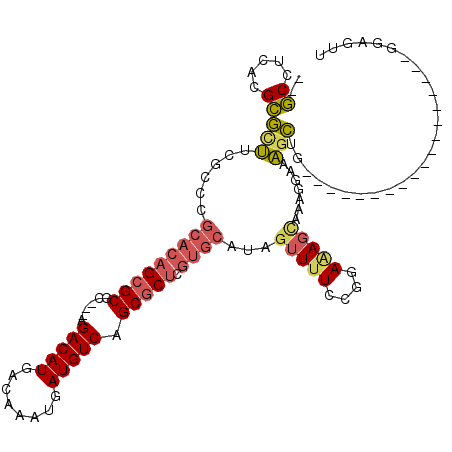
| Location | 9,467,657 – 9,467,751 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
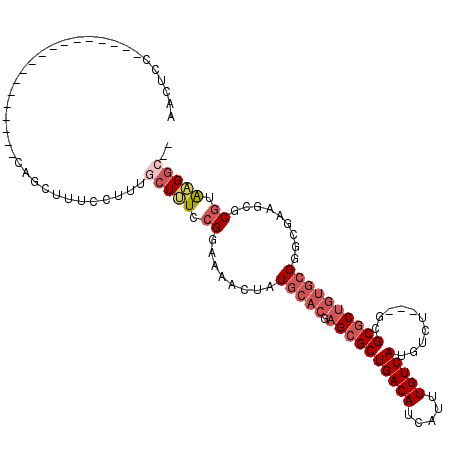
>2L_DroMel_CAF1 9467657 94 - 22407834 --GCCUUACGCGCUUCGUCCGCACAGCGCGC---AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUUCCGGAAAGCAAAGCAAAGCUG------------------CGAGUU --......(((((((.((..(((((((((..---.(((((........))))).))))).))))...(((((....)))))...)).)))).)------------------)).... ( -32.10) >DroPse_CAF1 31709 104 - 1 --UCCACCAGAGCCAGGACCC--GAGCGCGAAGAAGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUU-UCCGGAUACUACUCGUAGACGGGGA-----UGGGG--AUGGGGAU- --(((.(((...((......(--((((((......(((((........))))).)))))))......((.-.(((..(((.....)))..)))..)-----)..))--.)))))).- ( -37.70) >DroSec_CAF1 32411 94 - 1 --GCCUCACGCGCUUCGCCCGCACAGCGCGC---AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUUCCGGAAAGCAAAGGAAAGCUG------------------UGAGUU --..((((((((...)))..(((((((((..---.(((((........))))).))))).))))..((((((((..........)))))))))------------------)))).. ( -37.00) >DroSim_CAF1 32263 94 - 1 --GCCUUACGCGCUUCGCCCGCACAGCGCGC---AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUUCCGGAAAGCAAAGGAAAGCUG------------------GGAGUU --((.....))(((((....(((((((((..---.(((((........))))).))))).)))).(((((((((..........)))))))))------------------))))). ( -35.40) >DroYak_CAF1 33120 102 - 1 --GCCUUACGCGCUUCGCCCGCACAGCGCAG---AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUU-CGGAGAGCAAAGGAAAGCCGGGA---------AAGCGAGGAGUU --..(((...(((((..((((((((((((..---.(((((........))))).))))).))))...(((((-(...........)))))).))).---------)))))..))).. ( -36.70) >DroAna_CAF1 31103 109 - 1 CUGCCUCACGCUUUUCGCCCGCACCGAGCGC---AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUUCCGGAAAGGAAAACACAGAGGG--GGAAACCGGA--CCGGGGGU- ..(((((...((((((.(((((((.((((((---.(((((........))))).))))))))))...(((((((.....))))))).....)))--.))))..)).--...)))))- ( -49.60) >consensus __GCCUCACGCGCUUCGCCCGCACAGCGCGC___AGACAUGACAAAUGAUGUCAGCGCUCGUGCAUAGUUUUCCGGAAAGCAAAGGAAAGCUG__________________GGAGUU ..((.....))(((......(((((((((......(((((........))))).))))).))))...(((((....))))).......))).......................... (-15.41 = -15.92 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:28 2006