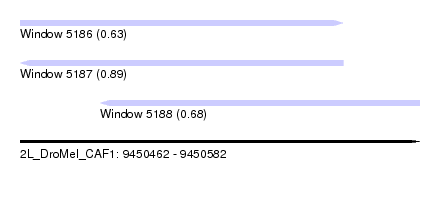
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,450,462 – 9,450,582 |
| Length | 120 |
| Max. P | 0.891562 |

| Location | 9,450,462 – 9,450,559 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
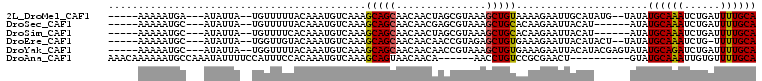
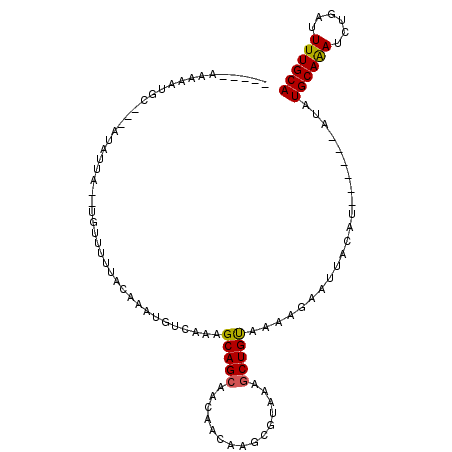
>2L_DroMel_CAF1 9450462 97 + 22407834 CAAGUCAAAACGAGUUUAUUGUGGUGCAAAAUCAGAUUUGCAUAUA--CAUAUGCAAUUCUUUUACAGCUUUACGCUAGUUGUUGCUGCUUUGACAUUU ...((((((((((.....)))).((((((....(((.(((((((..--..))))))).)))....(((((.......))))))))).)))))))).... ( -22.80) >DroSec_CAF1 14977 93 + 1 CAAGUUAAAACGAGUUUUUUGUUGUGCAAAAUCAGAUUUGCAUAU------AUGUAAUUCUUGUGCAGCUUUACGCUCGUUGUUGCUGCUUUGACAUUU ..(((...(((((((.....((((..((.....(((.(((((...------.))))).)))))..)))).....)))))))...)))............ ( -25.20) >DroSim_CAF1 14991 93 + 1 CAAGUCAAAACGAGUUUUUUGUUGUGCAAAAUCAGAUUUGCAUAU------AUGUAAUUCUUGUGCAGCUUUACGCUAGUUGUUGCUGCUUUGACAUUU ...(((((((((((..((....((((((((......)))))))).------....))..)))))(((((...((....))....))))))))))).... ( -27.40) >DroEre_CAF1 15263 96 + 1 CAAGUCAAAACGAGUUUUUUGUUGUGCAAAA-CAGAUUUGCAUAUA--AGUAUGUAAUUCUUUCACAGCUCUACGGUUGUUGUUGCUGCUUUGACAUUU ...((((((((((........))))(((.((-((((.(((((((..--..))))))).))....((((((....))))))))))..))))))))).... ( -22.30) >DroYak_CAF1 15318 99 + 1 CAAGUCAAAACGGGUUUUUUGUUGUGCAAAAUCAGAUCUGCAUAUACUCGUAUGUAAUUCUUUCACAGCUUUACGGUUGUUGUUGCUGCUUUGACAUUU ...((((((.(((...(((((.....)))))..(((..(((((((....)))))))..)))...((((((....)))))).....))).)))))).... ( -21.90) >DroAna_CAF1 14509 83 + 1 CAGGUCAAAACUUGUUUUUUGUUGUGCAAAACACAAUUUGCAUAC----------AGUUCGCGGACAGGUU------UGUUGUUACUGCUUUGACAUUU ...((((((((((((((.((((.(((((((......)))))))))----------)).....)))))))))------)...((....))..)))).... ( -19.60) >consensus CAAGUCAAAACGAGUUUUUUGUUGUGCAAAAUCAGAUUUGCAUAU______AUGUAAUUCUUGUACAGCUUUACGCUUGUUGUUGCUGCUUUGACAUUU ...((((((.............((((((((......)))))))).....................((((...((....))....)))).)))))).... (-14.69 = -15.38 + 0.69)


| Location | 9,450,462 – 9,450,559 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9450462 97 - 22407834 AAAUGUCAAAGCAGCAACAACUAGCGUAAAGCUGUAAAAGAAUUGCAUAUG--UAUAUGCAAAUCUGAUUUUGCACCACAAUAAACUCGUUUUGACUUG ....((((((((..........(((.....)))((((((((.(((((((..--..))))))).)))...)))))..............))))))))... ( -23.90) >DroSec_CAF1 14977 93 - 1 AAAUGUCAAAGCAGCAACAACGAGCGUAAAGCUGCACAAGAAUUACAU------AUAUGCAAAUCUGAUUUUGCACAACAAAAAACUCGUUUUAACUUG ..................((((((.((((..((.....))..))))..------...((((((......))))))..........))))))........ ( -15.20) >DroSim_CAF1 14991 93 - 1 AAAUGUCAAAGCAGCAACAACUAGCGUAAAGCUGCACAAGAAUUACAU------AUAUGCAAAUCUGAUUUUGCACAACAAAAAACUCGUUUUGACUUG ....((((((((.((........))((((..((.....))..))))..------...((((((......)))))).............))))))))... ( -20.00) >DroEre_CAF1 15263 96 - 1 AAAUGUCAAAGCAGCAACAACAACCGUAGAGCUGUGAAAGAAUUACAUACU--UAUAUGCAAAUCUG-UUUUGCACAACAAAAAACUCGUUUUGACUUG ....((((((((.............((((..((.....))..)))).....--....((((((....-.)))))).............))))))))... ( -18.70) >DroYak_CAF1 15318 99 - 1 AAAUGUCAAAGCAGCAACAACAACCGUAAAGCUGUGAAAGAAUUACAUACGAGUAUAUGCAGAUCUGAUUUUGCACAACAAAAAACCCGUUUUGACUUG ....((((((((..........((((((....(((((.....))))))))).))...((((((......)))))).............))))))))... ( -20.50) >DroAna_CAF1 14509 83 - 1 AAAUGUCAAAGCAGUAACAACA------AACCUGUCCGCGAACU----------GUAUGCAAAUUGUGUUUUGCACAACAAAAAACAAGUUUUGACCUG ....((((((((.((.(((...------....)))..))....(----------((.((((((......))))))..)))........))))))))... ( -16.00) >consensus AAAUGUCAAAGCAGCAACAACAAGCGUAAAGCUGUAAAAGAAUUACAU______AUAUGCAAAUCUGAUUUUGCACAACAAAAAACUCGUUUUGACUUG ....((((((((.............((((..((.....))..))))...........((((((......)))))).............))))))))... (-15.76 = -16.15 + 0.39)



| Location | 9,450,486 – 9,450,582 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.93 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9450486 96 - 22407834 -----AAAAAUGA---AUAUUA--UGUUUUUACAAAUGUCAAAGCAGCAACAACUAGCGUAAAGCUGUAAAAGAAUUGCAUAUG--UAUAUGCAAAUCUGAUUUUGCA -----........---......--((((((.((....)).))))))((((....((((.....))))....(((.(((((((..--..))))))).)))....)))). ( -20.40) >DroSec_CAF1 15001 92 - 1 -----AAAAAUGC---AUAUUA--UGUUUUUACAAAUGUCAAAGCAGCAACAACGAGCGUAAAGCUGCACAAGAAUUACAU------AUAUGCAAAUCUGAUUUUGCA -----.....(((---((((.(--(((..((.....(((....(((((....((....))...))))))))..))..))))------))))))).............. ( -17.10) >DroSim_CAF1 15015 92 - 1 -----AAAAAUGC---AUAUUA--UGUUUUCACAAAUGUCAAAGCAGCAACAACUAGCGUAAAGCUGCACAAGAAUUACAU------AUAUGCAAAUCUGAUUUUGCA -----.....(((---((((.(--(((.(((((....))....(((((....((....))...)))))....)))..))))------))))))).............. ( -18.30) >DroEre_CAF1 15287 95 - 1 -----AAAAAUGC---AUAUUA--UGGUUGUACAAAUGUCAAAGCAGCAACAACAACCGUAGAGCUGUGAAAGAAUUACAUACU--UAUAUGCAAAUCUG-UUUUGCA -----.....(((---((((((--(((((((.....(((....)))......)))))))))(((.(((((.....)))))..))--))))))))......-....... ( -19.90) >DroYak_CAF1 15342 98 - 1 -----AAAAAUGC---AUAUUA--UGGUUUUACAAAUGUCAAAGCAGCAACAACAACCGUAAAGCUGUGAAAGAAUUACAUACGAGUAUAUGCAGAUCUGAUUUUGCA -----....((((---...(((--(((((.......(((....)))........))))))))...(((((.....))))).....)))).((((((......)))))) ( -16.36) >DroAna_CAF1 14533 92 - 1 AAACAAAAAAUGCCAAAUAUUUUCCAUUUCCACAAAUGUCAAAGCAGUAACAACA------AACCUGUCCGCGAACU----------GUAUGCAAAUUGUGUUUUGCA ...................(((..(((((....)))))..)))(((((....(((------....)))......)))----------)).((((((......)))))) ( -9.60) >consensus _____AAAAAUGC___AUAUUA__UGUUUUUACAAAUGUCAAAGCAGCAACAACAAGCGUAAAGCUGUAAAAGAAUUACAU______AUAUGCAAAUCUGAUUUUGCA ...........................................(((((...............)))))......................((((((......)))))) (-10.12 = -9.93 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:24 2006