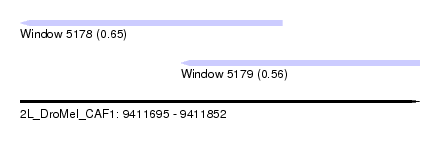
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,411,695 – 9,411,852 |
| Length | 157 |
| Max. P | 0.647085 |

| Location | 9,411,695 – 9,411,798 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
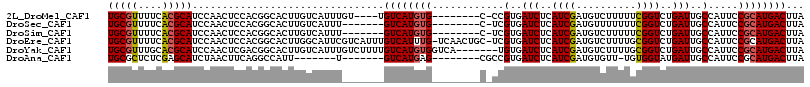
>2L_DroMel_CAF1 9411695 103 - 22407834 UGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUUGU----UGUCAUGUG--------C-CCGUGAUCUCAUCGAUGUCUUUUUCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))...........(((....)))......----.((((((((--------.-..(..(((..(((((........)))))..)))..).....))))))))... ( -28.00) >DroSec_CAF1 40272 100 - 1 UGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU-------GUCAUGUG--------C-UCGUGAUCUCAUCGAUGUUUUUUUCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))...........(((....)))....-------((((((((--------.-..(..(((..(((((........)))))..)))..).....))))))))... ( -28.00) >DroSim_CAF1 42822 100 - 1 UGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU-------GUCAUGUG--------C-UCGUGAUCUCAUCGAUGUCUUUUUCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))...........(((....)))....-------((((((((--------.-..(..(((..(((((........)))))..)))..).....))))))))... ( -28.00) >DroEre_CAF1 40711 114 - 1 UGCGUUUUCACGCAUCCAACUCCACGGCACUUGGCAUUCGUCAUUUGUCAUUUG-UCAACUGC-UCGUGAUCUCAUCGAUGUCUUUUGCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))........(((((((.((((((...(.(....).)...))-)))).)))-.))))...........(((...(((((..((.....))..))))).)))... ( -32.30) >DroYak_CAF1 42546 109 - 1 UGCGUUUGCACGCAUCCAACUCGACGGCACUUGUCAUUUGUCUUUUGUCAUGUGGUCA-------UGUGAUCUCAUCGAUGUCUUUUGCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))........((((((....)))....)))....(((((((((...-------.(..(((..((((..........))))..)))..)....)))))))))... ( -30.40) >DroAna_CAF1 43865 93 - 1 UGCGCUCUCGAGCAUCUAACUUCAGGCCAUU-------U-------GUCAUGAG--------CGCCGUGAUCUCAUCGAUGUGUU-UGUGGUAUGAUUGCCAUUCCGCAUGACUUA .((((((..(((........))).(((....-------.-------)))..)))--------)))((((....)).))(((((..-.((((((....))))))..)))))...... ( -23.80) >consensus UGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU_______GUCAUGUG________C_UCGUGAUCUCAUCGAUGUCUUUUGCGGUCUGAUUGCCAUUCCGCAUGACUUA (((((....)))))................................((((((((............(..(((..((((..........))))..)))..).....))))))))... (-17.94 = -18.03 + 0.09)
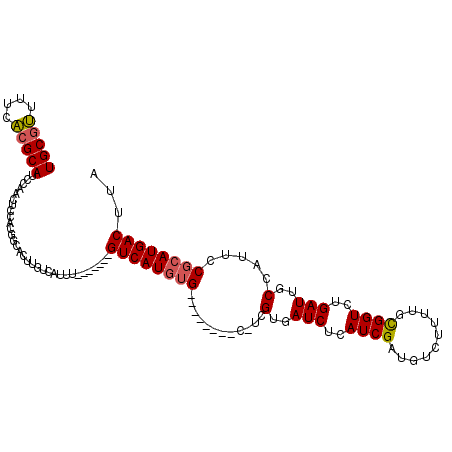
| Location | 9,411,758 – 9,411,852 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9411758 94 - 22407834 CACCACAUAACCGAGCUCGU---CUUAAGUAUGUGGACUCCUUGUCGUC-GCGAGAAAUGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUUG ...((((((...(((.....---)))...))))))(((....((((((.-(.(((..((((((....))))))....))))))))))...)))..... ( -25.00) >DroSec_CAF1 40333 93 - 1 CACCACAUAACCGAGUUCGU---CUUAAGUAUGUGGACUCCUUGUCGCU-CCGAGAAAUGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU- ...((((((...(((.....---)))...))))))(((....(((((..-..(((..((((((....))))))....)))..)))))...)))....- ( -23.00) >DroSim_CAF1 42883 93 - 1 CACCACAUAACCGAGUUCGU---CUUAAGUAUGUGGACUCCUUGUCGCU-CCGAGAAAUGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU- ...((((((...(((.....---)))...))))))(((....(((((..-..(((..((((((....))))))....)))..)))))...)))....- ( -23.00) >DroEre_CAF1 40785 94 - 1 CACCACAUAACCGAGCUCGU---CUUAAGUAUGUGGACUGCUUGUCGCC-CCGAGCAAUGCGUUUUCACGCAUCCAACUCCACGGCACUUGGCAUUCG ..(((((((...(((.....---)))...)))))))..((((.((.(((-..(((..((((((....))))))....)))...)))))..)))).... ( -26.90) >DroYak_CAF1 42615 94 - 1 CACCACAUAACCGAGUUCGC---CUUAAGUAUGUGGACUCCUUGUCGUC-CCGAGAAAUGCGUUUGCACGCAUCCAACUCGACGGCACUUGUCAUUUG ...((((((...(((.....---)))...))))))(((....(((((((-..(((..((((((....))))))....))))))))))...)))..... ( -27.30) >DroAna_CAF1 43926 90 - 1 CACCACAUAACCGAGUCAAUAUACAUAAGUAUGUGGAUGCCUUGUUGCCUCCGAGGAAUGCGCUCUCGAGCAUCUAACUUCAGGCCAUU-------U- ..(((((((.....((......)).....)))))))..((((.((((.(((.((((.......)))))))....))))...))))....-------.- ( -19.10) >consensus CACCACAUAACCGAGUUCGU___CUUAAGUAUGUGGACUCCUUGUCGCC_CCGAGAAAUGCGUUUUCACGCAUCCAACUCCACGGCACUUGUCAUUU_ ..(((((((...(((........)))...)))))))..........(((...(((..((((((....))))))....)))...)))............ (-19.90 = -19.60 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:15 2006