
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
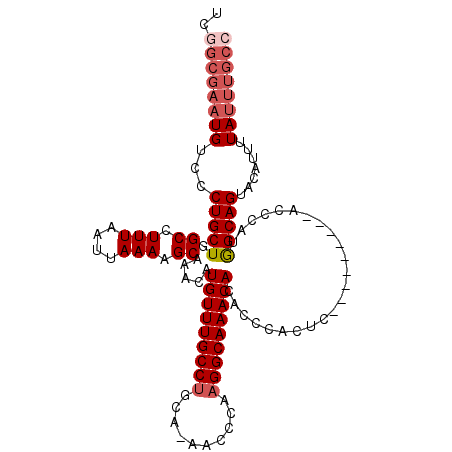
| Location | 9,411,125 – 9,411,227 |
| Length | 102 |
| Max. P | 0.987895 |

| Location | 9,411,125 – 9,411,227 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -22.14 |
| Energy contribution | -23.33 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
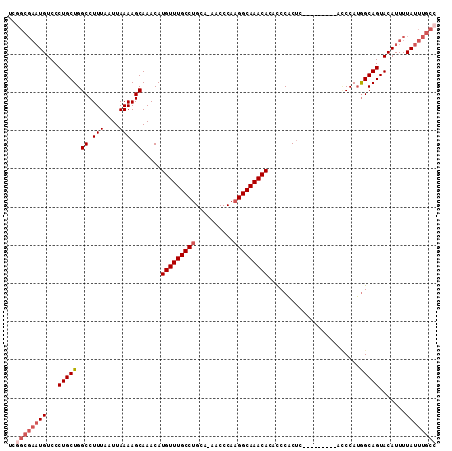
>2L_DroMel_CAF1 9411125 102 + 22407834 UCGGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA-AACCCAAGGCAAACACACCCACUC---------ACCCAUGGCAGUACAUUUUAUUUGCC ..((((((((....(((((.(((((.....))))......(((((((((...-......))))))))).........---------......).))))).....)))))))) ( -26.90) >DroSec_CAF1 39717 102 + 1 UCGGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA-AACCCAAGGCAAACACACCCACUC---------ACCCAUGGCAGUACAUUUUAUUUGCC ..((((((((....(((((.(((((.....))))......(((((((((...-......))))))))).........---------......).))))).....)))))))) ( -26.90) >DroSim_CAF1 42274 102 + 1 UCGGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA-AACCCAAGGCAAACACACCCACUC---------ACCCAUGGCAGUACAUUUUAUUUGCC ..((((((((....(((((.(((((.....))))......(((((((((...-......))))))))).........---------......).))))).....)))))))) ( -26.90) >DroEre_CAF1 40110 102 + 1 UCGGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA-AACCCAAGGCAAACACACCCACUC---------AGCCAUGGCAGUACAUUUUAUUUGCC ..((((((((...((((((((((((.....))))......(((((((((...-......))))))))).........---------.)))..))))).......)))))))) ( -29.50) >DroYak_CAF1 41914 102 + 1 UGUGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA-AACCCAAGGCAAACACACCCACUC---------ACCCAUGGCAGUACAUUUUAUUUGCC ...(((((((....(((((.(((((.....))))......(((((((((...-......))))))))).........---------......).))))).....))))))). ( -24.10) >DroAna_CAF1 42425 106 + 1 UC------UGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCGGCAGAACCCAAGGCAAACACAUCCCCCCCGACACUCCUUCCAGAGCAGUACAUUUUAUUUGCC ..------.....((((((((..........(((((....))))))))))))).......((((((....((......))..(((......)))............)))))) ( -20.30) >consensus UCGGCGAAUGUCCCUGCUGGCCUUUAAUUAAAAGCAAACAUGUUUGCCUGCA_AACCCAAGGCAAACACACCCACUC_________ACCCAUGGCAGUACAUUUUAUUUGCC ..((((((((...(((((.((.(((....))).)).....(((((((((..........)))))))))........................))))).......)))))))) (-22.14 = -23.33 + 1.19)

| Location | 9,411,125 – 9,411,227 |
|---|---|
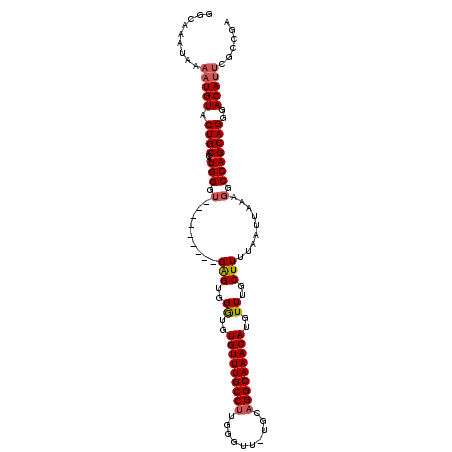
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
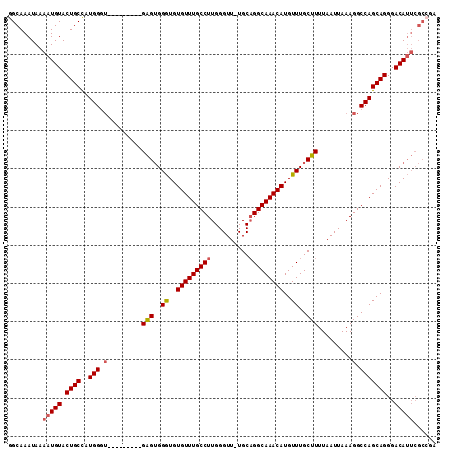
>2L_DroMel_CAF1 9411125 102 - 22407834 GGCAAAUAAAAUGUACUGCCAUGGGU---------GAGUGGGUGUGUUUGCCUUGGGUU-UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCCGA ((((............))))...(((---------(((((..(.((((.(((((..(((-.(((((((....)))))))....)))..))))).)))).)..)))))))).. ( -39.40) >DroSec_CAF1 39717 102 - 1 GGCAAAUAAAAUGUACUGCCAUGGGU---------GAGUGGGUGUGUUUGCCUUGGGUU-UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCCGA ((((............))))...(((---------(((((..(.((((.(((((..(((-.(((((((....)))))))....)))..))))).)))).)..)))))))).. ( -39.40) >DroSim_CAF1 42274 102 - 1 GGCAAAUAAAAUGUACUGCCAUGGGU---------GAGUGGGUGUGUUUGCCUUGGGUU-UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCCGA ((((............))))...(((---------(((((..(.((((.(((((..(((-.(((((((....)))))))....)))..))))).)))).)..)))))))).. ( -39.40) >DroEre_CAF1 40110 102 - 1 GGCAAAUAAAAUGUACUGCCAUGGCU---------GAGUGGGUGUGUUUGCCUUGGGUU-UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCCGA (((......(((((.((((..(((((---------(((..((..(((((((((.(....-..))))))))))..))..))).........)))))))))..))))).))).. ( -38.70) >DroYak_CAF1 41914 102 - 1 GGCAAAUAAAAUGUACUGCCAUGGGU---------GAGUGGGUGUGUUUGCCUUGGGUU-UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCACA ((((............))))....((---------(((((..(.((((.(((((..(((-.(((((((....)))))))....)))..))))).)))).)..)))))))... ( -36.10) >DroAna_CAF1 42425 106 - 1 GGCAAAUAAAAUGUACUGCUCUGGAAGGAGUGUCGGGGGGGAUGUGUUUGCCUUGGGUUCUGCCGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACA------GA ...........(((.(((((..((....(((...((((.(((((((((((((...((.....))))))))))))))).))))..))).....)))))))..)))------.. ( -31.40) >consensus GGCAAAUAAAAUGUACUGCCAUGGGU_________GAGUGGGUGUGUUUGCCUUGGGUU_UGCAGGCAAACAUGUUUGCUUUUAAUUAAAGGCCAGCAGGGACAUUCGCCGA .........(((((.((((..(((.(.........(((..((..(((((((((..........)))))))))..))..))).........).)))))))..)))))...... (-30.48 = -30.87 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:13 2006