| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 985,893 – 985,985 |
| Length | 92 |
| Max. P | 0.521112 |

| Location | 985,893 – 985,985 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -18.34 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
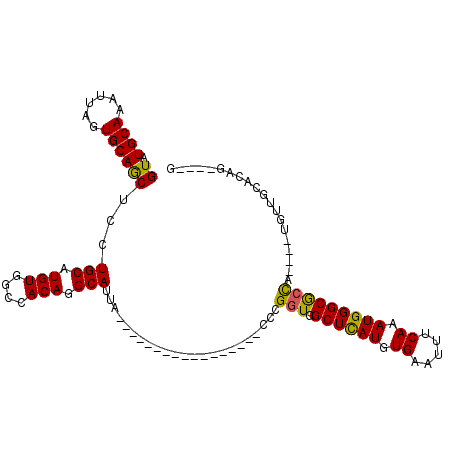
>2L_DroMel_CAF1 985893 92 - 22407834 GUAUGCAAAUUAGUGCAGCUCCUGCAUGUGGCCACAGGCAUUA-----------------CCCGGUGGCUCAUGUGAAUUUCAAAUGGGCGCCA----UGUUGCACAGGAGAG ...((((......)))).(((((((((((((((((.((.....-----------------.)).)))((((((.((.....)).))))))))))----)).)))..))))).. ( -33.40) >DroSec_CAF1 106799 92 - 1 GUAUGCAAAUUAGUGCAGCUCCUGCAUGUGGCCACAGGCAUUA-----------------CCCGGUGGCUCAUGUGAAUUUCAAAUGGGCGCCA----UGUUGCACAGGAGAG ...((((......)))).(((((((((((((((((.((.....-----------------.)).)))((((((.((.....)).))))))))))----)).)))..))))).. ( -33.40) >DroSim_CAF1 107406 92 - 1 GUAUGCAAAUUAGUGCAGCUCCUGCAUGUGGCCACAGGCAUUA-----------------CCCGGUGGCUCAUGUGAAUUUCAAAUGGGCGCCA----UGUUGCACAGGAGAG ...((((......)))).(((((((((((((((((.((.....-----------------.)).)))((((((.((.....)).))))))))))----)).)))..))))).. ( -33.40) >DroEre_CAF1 112952 82 - 1 GUAUGCAAAUUAGUGCAGCUCCUGCCUGUGGCCACAGGCAUUA-----------------CCCGGUGGCUUAUGUGAAUUUCAAAUGGGCGCUUCUUCU-------------- ...((((......))))((...(((((((....)))))))...-----------------.......((((((.((.....)).)))))))).......-------------- ( -22.50) >DroYak_CAF1 113187 90 - 1 GUAUGCAAAUUAGUGCAGCUCCUGCAUGUGGCCACAGGCAUUA-----------------CCCGCUGGCUCAUGUGAAUUUCAAAUGGGCGCCUCUUCUGUUUCUU------C ....(((.....((((((...))))))(.(((....(((....-----------------...))).((((((.((.....)).))))))))).)...))).....------. ( -21.20) >DroAna_CAF1 101109 103 - 1 GUAUGCAAAUUAGUGCAACUCCUGCAUGUGGCUACAGGCAUUACACCACCACCCCCAUCACCCGGU-GCUCAUGUGAAUUUCAAAUGGGCACCA----UGUUCCCCCG----- ((((((......(((((.....))))).((....)).))).)))...................(((-((((((.((.....)).))))))))).----..........----- ( -25.60) >consensus GUAUGCAAAUUAGUGCAGCUCCUGCAUGUGGCCACAGGCAUUA_________________CCCGGUGGCUCAUGUGAAUUUCAAAUGGGCGCCA____UGUUGCACAG____G ((.((((......))))))...(((.(((....))).))).......................(((.((((((.((.....)).))))))))).................... (-18.34 = -17.95 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:04 2006