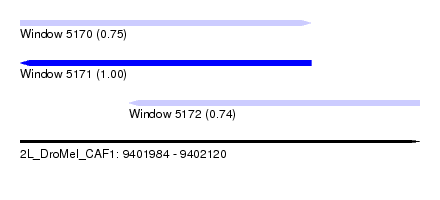
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,401,984 – 9,402,120 |
| Length | 136 |
| Max. P | 0.999258 |

| Location | 9,401,984 – 9,402,083 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9401984 99 + 22407834 ACGAGCAUUUGCAUUUUACUUCUGCCUUCACUUAUUUUU-UUUCACAGCUGACUUUCU---UUGCUUUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..(((((((((((.......((.((..............-.......)).))......---.)))...........(((.(((.....))).))))))))))) ( -16.74) >DroSec_CAF1 30684 97 + 1 ACGAGCAUUUGCAUUUCACUUCUGCCUUCACUUAUUCUU-UUUCACAACUGACUUUCU---UGC--UUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..((((((((((((((.....((((...(((........-..................---...--.........)))..)))).))))))....)))))))) ( -15.26) >DroSim_CAF1 32848 97 + 1 ACGAGCAUUUGCAUUUCACUUCUGCCUUCACUUAUUCUU-UUUCACAGCUGACUUUUU---UAG--UUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..(((((((..............................-...(((((..((((....---.))--)).....)))))((((.((....)).))))))))))) ( -18.10) >DroEre_CAF1 31201 101 + 1 ACGAGCAUUUGCAUUUUACUUCUGCCUUCACUUAUUCUUUUUUCACAACCGACUUUUUGUGUUU--UUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..((((((((((((((.....((((...(((............(((((........)))))...--.........)))..)))).))))))....)))))))) ( -21.05) >DroYak_CAF1 32695 97 + 1 ACGAACAUUUGCAUUUCACUUCUGCCUUCACUUAUUCUU-UUUCACAACUGAGUUUCU---UGC--UUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..........(((((........................-...(((((..((((....---.))--)).....)))))((((.((....)).))))))))).. ( -16.70) >consensus ACGAGCAUUUGCAUUUCACUUCUGCCUUCACUUAUUCUU_UUUCACAACUGACUUUCU___UGC__UUUUUUAUUGUGCUGCAGCAAAUGCCGCAGAAUGCUU ..((((((((((((((.....((((...(((............................................)))..)))).))))))....)))))))) (-13.80 = -14.00 + 0.20)

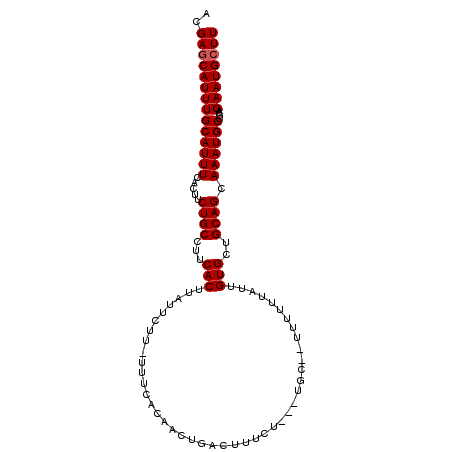

| Location | 9,401,984 – 9,402,083 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9401984 99 - 22407834 AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAAAGCAA---AGAAAGUCAGCUGUGAAA-AAAAAUAAGUGAAGGCAGAAGUAAAAUGCAAAUGCUCGU .(((((((((((((....))))))).............(((.---......((.(((......-.............))).)).......))).))))))... ( -20.65) >DroSec_CAF1 30684 97 - 1 AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAA--GCA---AGAAAGUCAGUUGUGAAA-AAGAAUAAGUGAAGGCAGAAGUGAAAUGCAAAUGCUCGU .((((((.((((.(((((.((((.((...........--)).---......(((.((((....-....)))).)))..)))))))))...))))))))))... ( -25.30) >DroSim_CAF1 32848 97 - 1 AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAA--CUA---AAAAAGUCAGCUGUGAAA-AAGAAUAAGUGAAGGCAGAAGUGAAAUGCAAAUGCUCGU .((((((.((((.(((((.((((..(((........(--((.---....)))...((......-.)).....)))...)))))))))...))))))))))... ( -23.00) >DroEre_CAF1 31201 101 - 1 AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAA--AAACACAAAAAGUCGGUUGUGAAAAAAGAAUAAGUGAAGGCAGAAGUAAAAUGCAAAUGCUCGU .((((((.....((((((.((((..(((.........--...(((((........)))))............)))...)))).....)))))).))))))... ( -22.15) >DroYak_CAF1 32695 97 - 1 AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAA--GCA---AGAAACUCAGUUGUGAAA-AAGAAUAAGUGAAGGCAGAAGUGAAAUGCAAAUGUUCGU .((((((.((((.(((((.((((.((...........--)).---......(((.((((....-....)))).)))..)))))))))...))))))))))... ( -22.80) >consensus AAGCAUUCUGCGGCAUUUGCUGCAGCACAAUAAAAAA__CAA___AGAAAGUCAGUUGUGAAA_AAGAAUAAGUGAAGGCAGAAGUGAAAUGCAAAUGCUCGU .(((((((((((((....)))))))..........................(((.((((.........)))).)))..(((.........))).))))))... (-19.28 = -19.36 + 0.08)

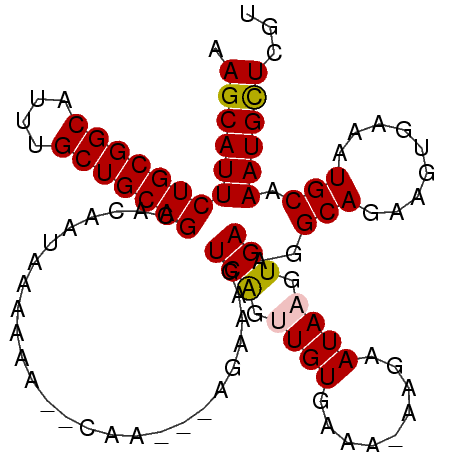
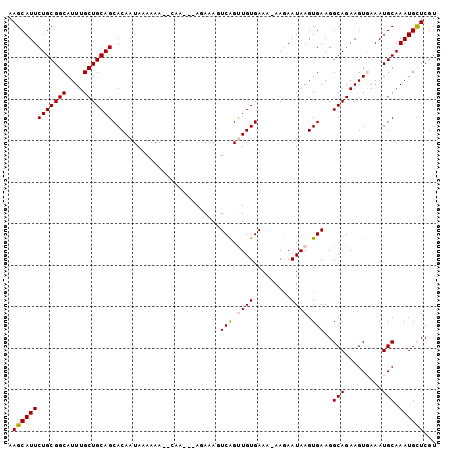
| Location | 9,402,021 – 9,402,120 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9402021 99 - 22407834 AGUGGAAG--GGCGCUGUGGUGGAUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA--AUAAAAAAAGCAA----AGAAAGUCA--GCUGUGAAA-AA ......(.--(((((((..(..((((.((...((((....))))...))...))))..)..))))...--.............----.(.....).--))).)....-.. ( -25.70) >DroSec_CAF1 30721 97 - 1 AGGGGAAG--GGCGCUGUGGAGGAUGACAAAAUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA--AUAAAAAA--GCA----AGAAAGUCA--GUUGUGAAA-AA ........--...((((..(..((((.((.((((((....)))))).))...))))..)..))))(((--((......--...----.........--)))))....-.. ( -23.47) >DroSim_CAF1 32885 97 - 1 AGUGGAAG--GGCGCUGUGGAGGAUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA--AUAAAAAA--CUA----AAAAAGUCA--GCUGUGAAA-AA ((((....--..))))(..(....((((....((((....))))..(((((((....)))))))....--........--...----.....))))--.)..)....-.. ( -22.80) >DroEre_CAF1 31238 103 - 1 AGUGGAGGAUGACGCUGUGGAGGAUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA--AUAAAAAA--AAA-CACAAAAAGUCG--GUUGUGAAAAAA ((((........))))(((.............((((....))))..(((((((....)))))))))).--........--...-(((((.......--.)))))...... ( -24.40) >DroYak_CAF1 32732 97 - 1 AGUGGAAG--GACGCUGUGGAGGAUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA--AUAAAAAA--GCA----AGAAACUCA--GUUGUGAAA-AA ........--..(((....(((..........((((....))))..(((((((....)))))))....--........--...----.....))).--...)))...-.. ( -21.60) >DroAna_CAF1 33068 97 - 1 -------G--GACUUUGGUGAGGUUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUACUGCAGCAAACAAAAAAAAA--GAAACA-UGAAACCGAAACCAGAAAAA-AA -------.--...((((((..((((..((...((((....))))..((((((......))))))..............--......-)).))))...))))))....-.. ( -22.80) >consensus AGUGGAAG__GACGCUGUGGAGGAUGACAAAUUGCUGACAAGCAUUCUGCGGCAUUUGCUGCAGCACA__AUAAAAAA__GAA____AGAAAGUCA__GCUGUGAAA_AA .............((((..(..((((.((...((((....))))...))...))))..)..))))............................................. (-17.62 = -17.82 + 0.20)

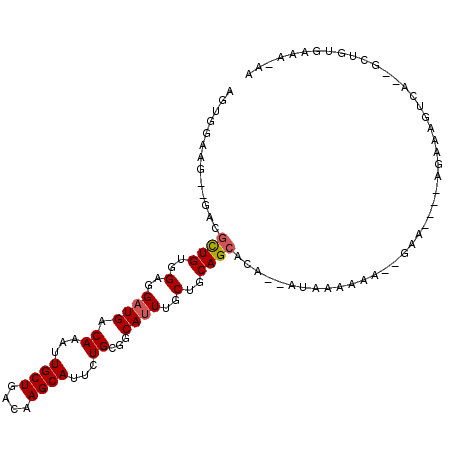
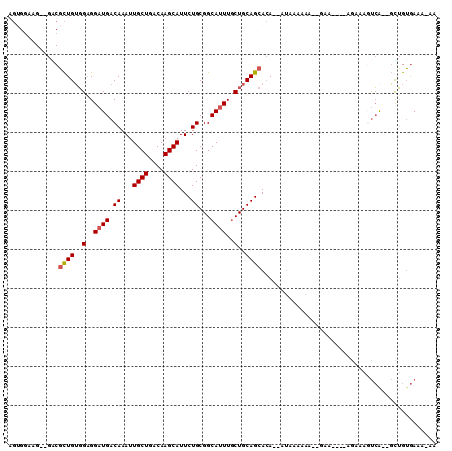
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:09 2006