
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,399,877 – 9,400,016 |
| Length | 139 |
| Max. P | 0.808475 |

| Location | 9,399,877 – 9,399,984 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -12.89 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
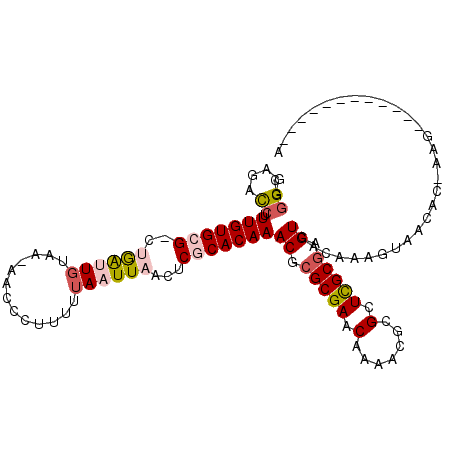
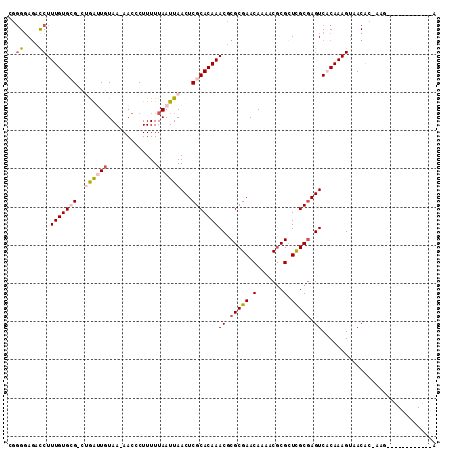
>2L_DroMel_CAF1 9399877 107 + 22407834 CGGGGAGACCUUUGUGCG-CUGAUUGUAA-AACCCUUUUUAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAAA-GCCCCAGCC-A .((((......(((((((-.((((((.((-(....))).))))))...)))))))((((((((((.......)))))))))).............-....-.))))....-. ( -35.00) >DroPse_CAF1 32554 99 + 1 GUGGCAGUCCUUUGUGGGGCAAGAUGUAAGAACCCUUUUUAAUUAACUCGCACAAACGCGCGAACAAAACGCGCUCGCGAGUCCCAAAGUAACAA-CAG------------A .(((..(((((....))))).........................((((((......(((((.......)))))..)))))).))).........-...------------. ( -23.40) >DroEre_CAF1 29053 94 + 1 CGGGGAGACCUUUGUGCG-CUGAUUGUAA-AACCCUUUUUAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-A--------------- .(((....)))(((((((-.((((((.((-(....))).))))))...)))))))((((((((((.......)))))))))).............-.--------------- ( -31.50) >DroYak_CAF1 30405 107 + 1 CGUGGAGACCUUUGUGCG-CUGAUUGUAA-AACCCUUUUUAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAGG-GCCGCAGCC-A .((((...((((((((((-.((((((.((-(....))).))))))...)))))).((((((((((.......)))))))))).............-))))-.))))....-. ( -35.80) >DroAna_CAF1 31337 110 + 1 GCGGGACCUCUUUGUGCG-CUGGUUCUAA-AACCCUUUUUAAUUAACUCGCACAAACGCGCGAACAAAACACGCUUGCCAGUCACAAAGUAACGGCAAGCGGCGCGAGUCGA ....(((...((((((((-..((((.(((-((....)))))...))))))))))))((((((.......).((((((((.((.((...)).))))))))))))))).))).. ( -35.70) >DroPer_CAF1 34920 99 + 1 GUGGGAGUCCUUUGUGGGGCAAGAUGUAAGAACCCUUUUUAAUUAACUCGCACAAACGCGCGAACAAAACGCGCUCGCGAGUCCCAAAGUAACAA-CAG------------A .((((((((((....)))))..........................(((((......(((((.......)))))..)))))))))).........-...------------. ( -27.90) >consensus CGGGGAGACCUUUGUGCG_CUGAUUGUAA_AACCCUUUUUAAUUAACUCGCACAAACGCGCGAACAAAACGCGCUCGCGAGUCACAAAGUAACAC_AAG____________A ..((....)).(((((((..((((((.............))))))...)))))))((.(((((.(.......).))))).)).............................. (-12.89 = -13.64 + 0.75)

| Location | 9,399,877 – 9,399,984 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
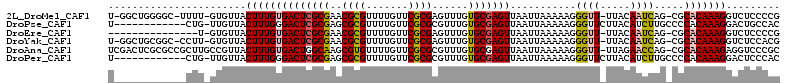
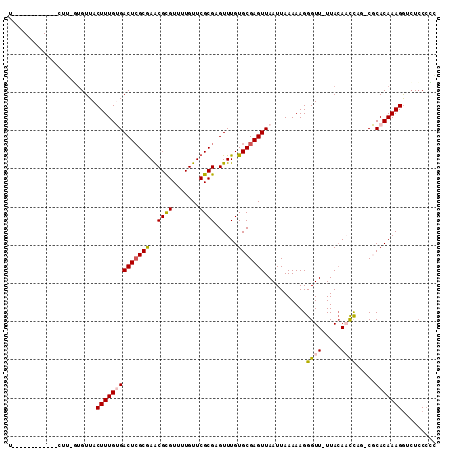
>2L_DroMel_CAF1 9399877 107 - 22407834 U-GGCUGGGGC-UUUU-GUGUUACUUUGUGACUCGCGAACGCGUUUUGUUCGCGAGUUUGUGCGAGUUAAUUAAAAAGGGUU-UUACAAUCAG-CGCACAAAGGUCUCCCCG .-((..(((((-((((-((((...(((((((((((((((((.....))))))))))))...)))))............((((-....))))..-.))))))))))))).)). ( -38.60) >DroPse_CAF1 32554 99 - 1 U------------CUG-UUGUUACUUUGGGACUCGCGAGCGCGUUUUGUUCGCGCGUUUGUGCGAGUUAAUUAAAAAGGGUUCUUACAUCUUGCCCCACAAAGGACUGCCAC .------------..(-(.(((.(((((.((((((((.(((((.......))))).....)))))))).........((((...........))))..)))))))).))... ( -31.40) >DroEre_CAF1 29053 94 - 1 ---------------U-GUGUUACUUUGUGACUCGCGAACGCGUUUUGUUCGCGAGUUUGUGCGAGUUAAUUAAAAAGGGUU-UUACAAUCAG-CGCACAAAGGUCUCCCCG ---------------.-......((((((((((((((((((.....)))))))))))).((((..(((...(((((....))-))).)))..)-)))))))))......... ( -28.70) >DroYak_CAF1 30405 107 - 1 U-GGCUGCGGC-CCUU-GUGUUACUUUGUGACUCGCGAACGCGUUUUGUUCGCGAGUUUGUGCGAGUUAAUUAAAAAGGGUU-UUACAAUCAG-CGCACAAAGGUCUCCACG .-(((....))-)...-(((..(((((..((((((((((((.....))))))))))))((((((.......(((((....))-))).......-)))))))))))...))). ( -34.84) >DroAna_CAF1 31337 110 - 1 UCGACUCGCGCCGCUUGCCGUUACUUUGUGACUGGCAAGCGUGUUUUGUUCGCGCGUUUGUGCGAGUUAAUUAAAAAGGGUU-UUAGAACCAG-CGCACAAAGAGGUCCCGC ..((((((((((((((((((((((...))))).))))))))..........)))))((((((((..............((((-....))))..-))))))))..)))).... ( -39.69) >DroPer_CAF1 34920 99 - 1 U------------CUG-UUGUUACUUUGGGACUCGCGAGCGCGUUUUGUUCGCGCGUUUGUGCGAGUUAAUUAAAAAGGGUUCUUACAUCUUGCCCCACAAAGGACUCCCAC .------------...-..(((.(((((.((((((((.(((((.......))))).....)))))))).........((((...........))))..))))))))...... ( -30.40) >consensus U____________CUU_GUGUUACUUUGUGACUCGCGAACGCGUUUUGUUCGCGAGUUUGUGCGAGUUAAUUAAAAAGGGUU_UUACAACCAG_CGCACAAAGGUCUCCCCC .......................((((((((((((((..((((.......))))......)))))))...........((((.....)))).....)))))))......... (-19.32 = -19.40 + 0.08)

| Location | 9,399,914 – 9,400,016 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9399914 102 + 22407834 UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAAA-GCCCCAGCC-AAAAACGCGUUUGCAAGCCAUUUUUAUUUGGCG .........(((....((((((((((.......)))))))))).............-....-((....)).-......))).......((((........)))). ( -25.80) >DroSec_CAF1 28660 101 + 1 UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAAA-GCCCCAGCC-AA-AACGCGUUUGCAAGCCAUUUUUAUUUGGCG ..........(((...((((((((((.......)))))))))).............-....-......((.-..-...))...)))..((((........)))). ( -26.00) >DroSim_CAF1 30766 101 + 1 UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAAA-GCCCCAGCC-AA-AACGCGUUUGCAAGCCAUUUUUAUUUGGCG ..........(((...((((((((((.......)))))))))).............-....-......((.-..-...))...)))..((((........)))). ( -26.00) >DroEre_CAF1 29090 87 + 1 UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-A-----------------AACGCGUUUGCAAGCCAUUUUUAUUUGGCG .........(((....((((((((((.......))))))))))......((.....-.-----------------.))))).......((((........)))). ( -24.70) >DroYak_CAF1 30442 101 + 1 UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC-AAGG-GCCGCAGCC-AA-AACGCGUUUGCAAGCCAUUUUUAUUUGGCG .........(((....((((((((((.......)))))))))).............-...(-((....)))-..-...))).......((((........)))). ( -29.80) >DroAna_CAF1 31374 104 + 1 UAAUUAACUCGCACAAACGCGCGAACAAAACACGCUUGCCAGUCACAAAGUAACGGCAAGCGGCGCGAGUCGAA-AACGCGUUUGCCAACCAUUUUUAUUUGGCG ..............(((((((............(((((((.((.((...)).)))))))))(((....)))...-..)))))))(((((..........))))). ( -32.00) >consensus UAAUUAACUCGCACAAACUCGCGAACAAAACGCGUUCGCGAGUCACAAAGUAACAC_AAAA_GCCCCAGCC_AA_AACGCGUUUGCAAGCCAUUUUUAUUUGGCG .........(((....((((((((((.......))))))))))...................((....))........))).......((((........)))). (-21.50 = -22.00 + 0.50)

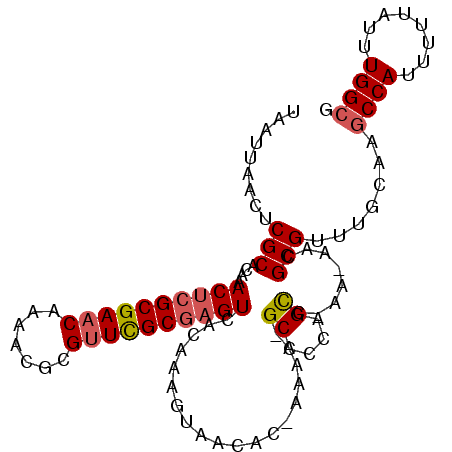
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:06 2006