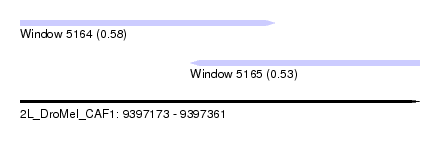
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,397,173 – 9,397,361 |
| Length | 188 |
| Max. P | 0.578997 |

| Location | 9,397,173 – 9,397,293 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -37.86 |
| Energy contribution | -37.06 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9397173 120 + 22407834 UCUCUACUUGGCAAACGGCUUCUUGGUGACCACAUUGGCUGCUUUCUUCUCCGCUUCACGCCUAGCAGCGGUUUCUUCGCGCUGCUUCCGGAUCAGAGCCAGCCUGGCCAAGUCGGCCCU ..((.(((((((....(((........).)).....(((((..((((..(((((.....))..((((((((......).)))))))...)))..)))).)))))..))))))).)).... ( -38.90) >DroSec_CAF1 25976 120 + 1 UCUCUACUUGGCAAACGGCUUCUUGGUGACCACAUUGGCUGCUUUCUUCUCCGCUUCACGCCUGGCAGCGGUCUCUUCGCGCUGCUUCCGGAUCAGAGCCAGCCUGGCCAAGUCGGCCAU ................((((.(((((...(((....(((((..((((..(((((.....))..((((((((......).)))))))...)))..)))).)))))))))))))..)))).. ( -39.60) >DroSim_CAF1 28051 120 + 1 UCUCUACUUGGCAAAAGGCUUCUUGGUGACCACAUUGGCUGCCUUCUUCUCCGCUUCACGCCUGGCAGCGGUCUCUUCGCGCUGCUUUCGGAUCAGAGCCAGCCUGGCCAAGUCGGCCAU ..((.(((((((...(((((....(((..((.....))..))).........((((....((.((((((((......).)))))))...))....)))).))))).))))))).)).... ( -43.40) >DroEre_CAF1 26374 120 + 1 CCUCUACUUGGCAAACGGCUUCUUGGUGACCGCAACGGCGGCCUUCUUCUCCGCCUCCCGCCUGGCGGCGGUUUCCUCGCGCUGCUUCCGGAUGAGAGCCAGCCUGGCCAAGUCGGCCCU ..((.(((((((....((((....((.((((((....))))......)).))((((((((...((((((((......).)))))))..)))..))).)).))))..))))))).)).... ( -43.90) >DroYak_CAF1 27663 120 + 1 UCUCUACUUGGCAAACGGCUUCUUGGUGACCAUAUCGGCUGCCUUCUUGUCCGCCGCCCGCUUGGCGGCGGUUUCCUCACGUUGCUUCCGGAUCAGAGCCAAUCUGGCCAAGUCGGCCCU ..((.(((((((....(((.....(((..((.....))..))).(((.((((((((((.....))))))((................)))))).))))))......))))))).)).... ( -42.09) >consensus UCUCUACUUGGCAAACGGCUUCUUGGUGACCACAUUGGCUGCCUUCUUCUCCGCUUCACGCCUGGCAGCGGUUUCUUCGCGCUGCUUCCGGAUCAGAGCCAGCCUGGCCAAGUCGGCCCU ..((.(((((((....(((.....(((..((.....))..))).(((..(((((.....))..((((((((......).)))))))...)))..))))))......))))))).)).... (-37.86 = -37.06 + -0.80)

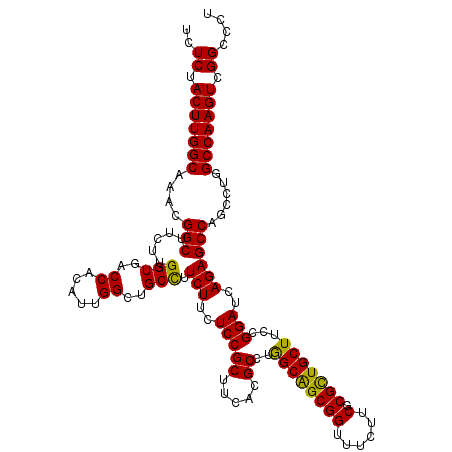
| Location | 9,397,253 – 9,397,361 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -31.22 |
| Energy contribution | -31.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9397253 108 - 22407834 GCGAUCAA------------GAUCAGAGUGCUCGCAAGCGUUACGAGAAACUCCAUGUAGCUGGGAAAACCACCGAGGCUAGGGCCGACUUGGCCAGGCUGGCUCUGAUCCGGAAGCAGC ((..((..------------(((((((((.((((.....((((((.(......).))))))(((.....))).))))(((..((((.....)))).)))..)))))))))..)).))... ( -39.70) >DroSec_CAF1 26056 120 - 1 GUGAUCGAGAGCAGGACAAGGAUCAGAGUGCUCGCAAGCGUUACGAGAAACUCCACGUCGCUGGAAAAACCACCGAGGCUAUGGCCGACUUGGCCAGGCUGGCUCUGAUCCGGAAGCAGC ..........((.......((((((((((.((((..((((..(((.(......).)))))))((.....))..))))(((.(((((.....))))))))..))))))))))....))... ( -40.20) >DroSim_CAF1 28131 120 - 1 GUGAUCGAGAGCAGGACAAGGAUCAGAGUGCUCGCAAGCGUUACGAGAAACUCCACGUCGCUGGAAAAACCACCGAGGCUAUGGCCGACUUGGCCAGGCUGGCUCUGAUCCGAAAGCAGC ..........((.......((((((((((.((((..((((..(((.(......).)))))))((.....))..))))(((.(((((.....))))))))..))))))))))....))... ( -40.20) >DroEre_CAF1 26454 120 - 1 ACGGCCGAGAUCAGCCCAAGGAGCAGAGUGGGCGCCAGCGCUAUGAGAAGCUCCAGUCCGCUGGCAAGACCACCGAGGCUAGGGCCGACUUGGCCAGGCUGGCUCUCAUCCGGAAGCAGC ..(((((((.((.((((..(((((......((((....)))).......)))))((((((.(((.....))).)).)))).)))).)))))))))..((((.((..(....)..)))))) ( -54.02) >DroYak_CAF1 27743 120 - 1 GCGGCCGAGAUUCGGCCAAGGAUCAGAGUGCACGUCAGCGUUAUGAGAAACUCCAUACCGCUGGUAAAACCACCGAGGCUAGGGCCGACUUGGCCAGAUUGGCUCUGAUCCGGAAGCAAC (((((((.....)))))..((((((((((.((.(((((((.((((.(.....))))).))))(((......)))..(((((((.....))))))).)))))))))))))))....))... ( -54.00) >consensus GCGAUCGAGAGCAGGACAAGGAUCAGAGUGCUCGCAAGCGUUACGAGAAACUCCACGUCGCUGGAAAAACCACCGAGGCUAGGGCCGACUUGGCCAGGCUGGCUCUGAUCCGGAAGCAGC ...................(((((((((...((.(.((((....(((...))).....)))))))....(((((..(((((((.....))))))).)).))))))))))))......... (-31.22 = -31.82 + 0.60)

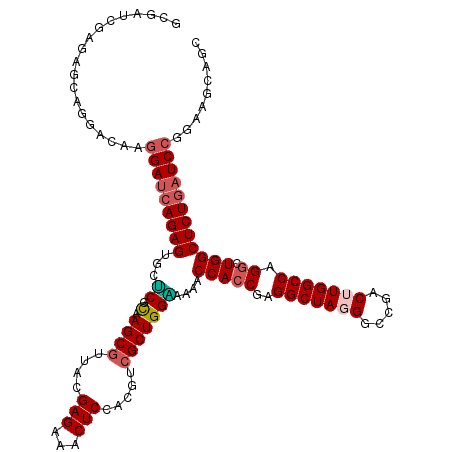
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:02 2006