
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,390,203 – 9,390,312 |
| Length | 109 |
| Max. P | 0.563574 |

| Location | 9,390,203 – 9,390,312 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9390203 109 + 22407834 UAUAUAUACUUAUAUGCUUAU--CA---AAAUUUAAUUUUCCGACAGAGUCUGGAGACCCUUGACAACGGCAAGCCCUACAGCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ((((((....)))))).....--..---...................((((.(((((.((.((.....((((.((......)).))))......)).)).))))).)))).... ( -21.80) >DroVir_CAF1 25878 101 + 1 CCU-----------UUCUAAU--CACCUUGCCUAUAUUUUCAUUUAGAGUCUGGAAACCCUGGACAAUGGCAAGCCCUACAGCAUGGCCUACAAUGUGGAUUUGCCCACGGCCA ...-----------.......--...((((((.......((.....))(((..(.....)..)))...))))))..........(((((......((((......))))))))) ( -25.70) >DroSec_CAF1 21869 108 + 1 UAUAUAUACUUAUAUGCUUAU--CA---A-UUUAAAUUUUCCAACAGAGUCUGGAGACCCUCGACAACGGCAAGCCCUACAGCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ((((((....)))))).....--..---.-.................((((.(((((.((.((.....((((.((......)).))))......)).)).))))).)))).... ( -24.20) >DroSim_CAF1 23956 108 + 1 UAUAUAUACUUAUAUGCUAAU--CA---A-UUUAAAUUUUCCGACAGAGUCUGGAGACCCUCGACAACGGCAAGCCCUACAGCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ((((((....)))))).....--..---.-.................((((.(((((.((.((.....((((.((......)).))))......)).)).))))).)))).... ( -24.20) >DroEre_CAF1 22177 108 + 1 AAUGUAUAUUUAUAUGCUUAU--CA---A-UUUCAAUUUCCCGCCAGAGUCUGGAGACCCUCGACAACGGCAAGCCCUACAGCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ...(((((....)))))....--..---.-............((...((((.(((((.((.((.....((((.((......)).))))......)).)).))))).)))))).. ( -24.90) >DroYak_CAF1 23440 110 + 1 UAUACAUACUUAUAUUUUCAAUGAA---A-UUUAUAUUUCCCGACAGAGUCUGGAGACCCUCGACAACGGAAAGCCCUACGCCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ......................(((---(-(....))))).......((((.(((((.((.((.....(((..((.....))....))).....)).)).))))).)))).... ( -22.60) >consensus UAUAUAUACUUAUAUGCUUAU__CA___A_UUUAAAUUUUCCGACAGAGUCUGGAGACCCUCGACAACGGCAAGCCCUACAGCAUGUCCUACAACGUGGAUCUCCCGACUGCCA ...............................................((((.(((((.((.((.....((((.((......)).)).)).....)).)).))))).)))).... (-16.79 = -17.27 + 0.48)

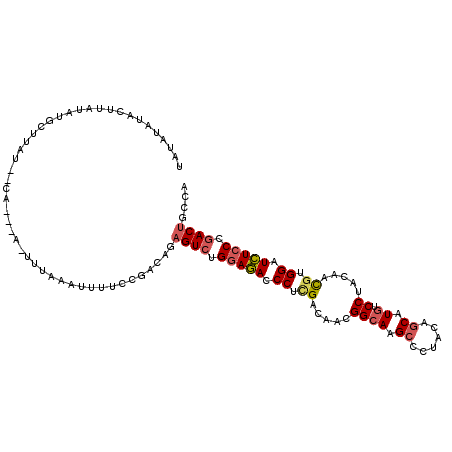
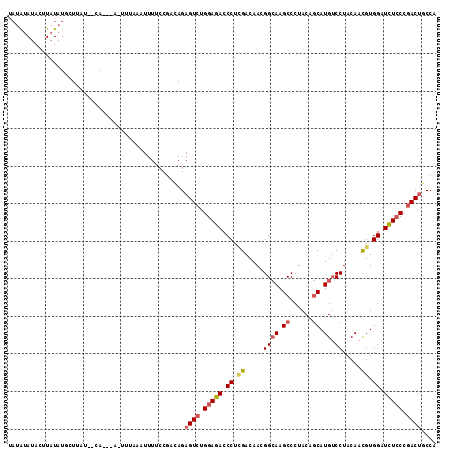
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:59 2006