
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,354,927 – 9,355,086 |
| Length | 159 |
| Max. P | 0.976586 |

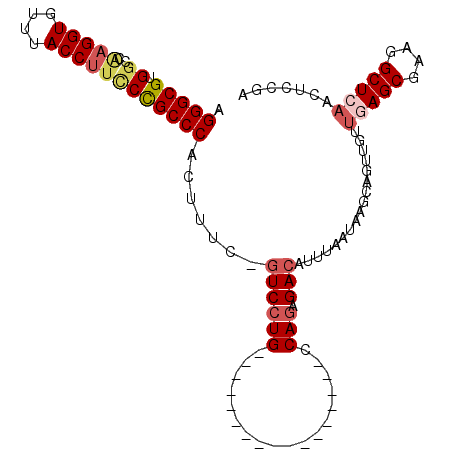
| Location | 9,354,927 – 9,355,017 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9354927 90 + 22407834 AGGGCGUGGCCAAGGUGUUUACCUUCCCGCCCACUUUG-GUCCUG-----------------CCAGAGACAUUUAAUAAGCAGUUGUUGAGCGAAGGCUCAACUCCGA .(((((.((..(((((....)))))))))))).(((((-(.....-----------------))))))..............(..(((((((....)))))))..).. ( -37.90) >DroSec_CAF1 23873 90 + 1 AGGGCGUGGCCAAGGUGUUUACCUUCCCGCCCACUUUG-GUCCUG-----------------CCAGAGACAUUUAAUAAGCAGUUGUUGAGCGAAGGCUCAACUCCGA .(((((.((..(((((....)))))))))))).(((((-(.....-----------------))))))..............(..(((((((....)))))))..).. ( -37.90) >DroSim_CAF1 23963 90 + 1 UGGGCGUGGCCAAGGUGUUUACCUUCCCGCCCACUUUC-GUCCUG-----------------CCAGAGACAUUUAAUAAGCAGUUGUUGAGCGAAGGCUCAACUCCGA ((((((.((..(((((....)))))))))))))...((-(..(((-----------------(................))))..(((((((....)))))))..))) ( -33.19) >DroEre_CAF1 24358 72 + 1 GGGGCGUGGCCGAGGUGUUUACCUUGCCGCCCACUUUC-GUCCUG-----------------CCAGAGACAUUUAAUAAGCAGUUGUUG------------------A .((((..(((.(((((....))))))))))))......-((((..-----------------...).)))..(((((((....))))))------------------) ( -19.70) >DroYak_CAF1 26372 90 + 1 AGGGCGUGGCCAAGGUGUUUACCUUUCCGCCCACUUUG-GUCCUG-----------------CCAGAGACAUUUAAUAAGCAGUUGCUGAGCGCAGGCUCAACACCGA .(((((.((..(((((....)))))))))))).(((((-(.....-----------------))))))..........(((....)))((((....))))........ ( -30.00) >DroAna_CAF1 36335 106 + 1 AGGGCGUGGCUGAGGUGCUUACCCGCCUGCCCACCUACCUUCCUGUCCAACUGGCAGGACGGGCAAGGACAUUUAAUAAGCAGC--CUAAGCGAAGGCUCAACACCCG .(((.((((((((((((......))))).........(((((((((((........))))))).))))............))))--)..(((....)))..)).))). ( -42.00) >consensus AGGGCGUGGCCAAGGUGUUUACCUUCCCGCCCACUUUC_GUCCUG_________________CCAGAGACAUUUAAUAAGCAGUUGUUGAGCGAAGGCUCAACUCCGA .(((((.((..(((((....)))))))))))).......((((((..................))).))).................(((((....)))))....... (-20.80 = -21.90 + 1.10)

| Location | 9,354,927 – 9,355,017 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.69 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9354927 90 - 22407834 UCGGAGUUGAGCCUUCGCUCAACAACUGCUUAUUAAAUGUCUCUGG-----------------CAGGAC-CAAAGUGGGCGGGAAGGUAAACACCUUGGCCACGCCCU ..((.(((((((....)))))))..(((((..............))-----------------)))..)-).....(((((((.((((....))))...)).))))). ( -34.44) >DroSec_CAF1 23873 90 - 1 UCGGAGUUGAGCCUUCGCUCAACAACUGCUUAUUAAAUGUCUCUGG-----------------CAGGAC-CAAAGUGGGCGGGAAGGUAAACACCUUGGCCACGCCCU ..((.(((((((....)))))))..(((((..............))-----------------)))..)-).....(((((((.((((....))))...)).))))). ( -34.44) >DroSim_CAF1 23963 90 - 1 UCGGAGUUGAGCCUUCGCUCAACAACUGCUUAUUAAAUGUCUCUGG-----------------CAGGAC-GAAAGUGGGCGGGAAGGUAAACACCUUGGCCACGCCCA .....(((((((....)))))))..(((((..............))-----------------))).((-....))(((((((.((((....))))...)).))))). ( -35.04) >DroEre_CAF1 24358 72 - 1 U------------------CAACAACUGCUUAUUAAAUGUCUCUGG-----------------CAGGAC-GAAAGUGGGCGGCAAGGUAAACACCUCGGCCACGCCCC .------------------......(((((..............))-----------------))).((-....))(((((((.((((....))))..)))..)))). ( -22.44) >DroYak_CAF1 26372 90 - 1 UCGGUGUUGAGCCUGCGCUCAGCAACUGCUUAUUAAAUGUCUCUGG-----------------CAGGAC-CAAAGUGGGCGGAAAGGUAAACACCUUGGCCACGCCCU ..((((((((((....)))))))..(((((..............))-----------------))).))-).....(((((..(((((....))))).....))))). ( -33.64) >DroAna_CAF1 36335 106 - 1 CGGGUGUUGAGCCUUCGCUUAG--GCUGCUUAUUAAAUGUCCUUGCCCGUCCUGCCAGUUGGACAGGAAGGUAGGUGGGCAGGCGGGUAAGCACCUCAGCCACGCCCU .(((((((((((....)))))(--((((..........((.((((((((.((((((......((......)).....)))))))))))))).))..))))))))))). ( -47.20) >consensus UCGGAGUUGAGCCUUCGCUCAACAACUGCUUAUUAAAUGUCUCUGG_________________CAGGAC_CAAAGUGGGCGGGAAGGUAAACACCUUGGCCACGCCCU .....(((((((....)))))))..(((((((((....(((.(((..................))))))....))))))))).(((((....)))))(((...))).. (-21.89 = -22.69 + 0.80)


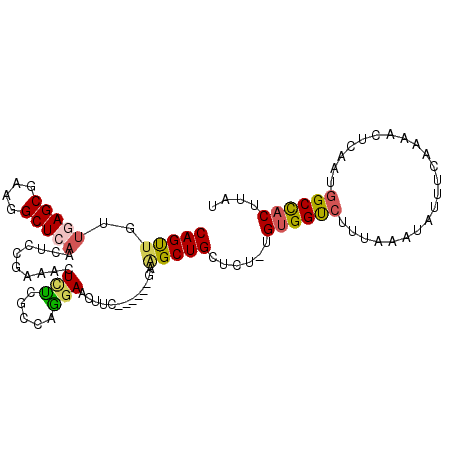
| Location | 9,354,989 – 9,355,086 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -13.87 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9354989 97 + 22407834 CAGUUGUUGAGCGAAGGCUCAACUCCGAAACUCUCACCAGGAAUUUC------GAAGCUGCUCU-UGUGGUCUUUAAAUAUUUCAAAACUCAAUAGCCACUUAU ((((((((((((....)))))))..(((((.(((.....))).))))------).)))))....-.(((((..((................))..))))).... ( -26.09) >DroSec_CAF1 23935 97 + 1 CAGUUGUUGAGCGAAGGCUCAACUCCGAAACUCUCGCCAGGAACUUC------GAAGCUGCUCU-UGUGGUCUUUAAAUAUUUCAAAACUCAAUGGCCACUUAU ((((((((((((....)))))))..((((..(((.....)))..)))------).)))))....-.((((((......................)))))).... ( -27.55) >DroSim_CAF1 24025 97 + 1 CAGUUGUUGAGCGAAGGCUCAACUCCGAAACUCUCGCCAGGAACUUC------GAAGCUGCUCU-UGUGGUCUUUAAAUAUUUCAAAACUCAAUGGCCACUUAU ((((((((((((....)))))))..((((..(((.....)))..)))------).)))))....-.((((((......................)))))).... ( -27.55) >DroEre_CAF1 24420 79 + 1 CAGUUGUUG------------------AAACUCCCGCCAGGAACUUC------GAAGCUGCUCU-UGUGGUCUUUAAAUAUUUCACAACUCAAUGGCCUCUUGU .((((((.(------------------(((.(((.....))).....------(((((..(...-.)..).)))).....)))))))))).............. ( -14.30) >DroYak_CAF1 26434 97 + 1 CAGUUGCUGAGCGCAGGCUCAACACCGAAACUUCGGCCAGGAACUUC------GAGGCUGCUCU-UGUGGUCUUUAAAUAUUUCAAAACUCAAUGGCCACUUAU ........(((.((((.(((....((((....))))....(.....)------))).)))).))-)((((((......................)))))).... ( -24.75) >DroAna_CAF1 36415 102 + 1 CAGC--CUAAGCGAAGGCUCAACACCCGGUUUCUGGGGAAAACUUUCCUCGCAGAAGCUGCACUUUGUGGCCUGUGCAUAUUUCAUAGCUCAACGGCUACUUAC ....--....(((.((((((((....((((((((((((((....)))).).)))))))))....))).))))).)))........(((((....)))))..... ( -31.70) >consensus CAGUUGUUGAGCGAAGGCUCAACUCCGAAACUCUCGCCAGGAACUUC______GAAGCUGCUCU_UGUGGUCUUUAAAUAUUUCAAAACUCAAUGGCCACUUAU (((((..(((((....)))))..........(((.....))).............)))))......((((((......................)))))).... (-13.87 = -14.17 + 0.30)

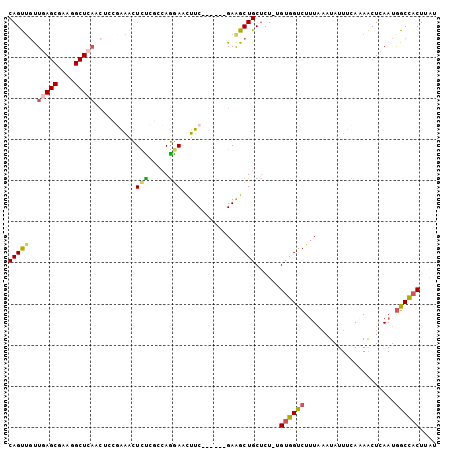
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:57 2006