
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,326,152 – 9,326,250 |
| Length | 98 |
| Max. P | 0.589801 |

| Location | 9,326,152 – 9,326,250 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.05 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
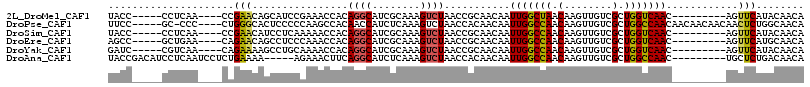
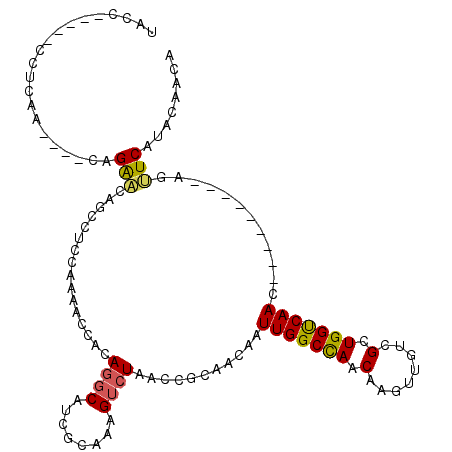
>2L_DroMel_CAF1 9326152 98 + 22407834 UACC-----CCUCAA----CCGAACAGCAUCCGAAACCACAGGCAUCGCAAAGUCUAACCGCAACAAUUGGCUAACAAGUUGUCGCUGGUCAAC---------AGUUCAUACAACA ....-----......----..((((.((((((.........)).)).))...((...(((((.((((((........)))))).)).)))..))---------.))))........ ( -17.00) >DroPse_CAF1 185019 106 + 1 UUCC-----GC-CCC----CUGGGCACUCCCCCAAGCCACAACCAUCUCAAAGUCUAACCACAACAAUUGGCCAACAAGUUGUCGCUGGCCAACAACAACAACAACUCUGGCAACA ....-----((-((.----..))))..........((((.............((......)).....(((((((.(........).)))))))...............)))).... ( -21.50) >DroSim_CAF1 159293 98 + 1 UACC-----CCUCAA----CCGAACAUCCUCAAAAACCACAGGCAUCGCAAAGUCUAACCGCAACAAUUGGCCAACAAGUUGUCGCUGGUCAAC---------AGUUCAUACAACA ....-----......----..((((...............((((........)))).(((((.((((((........)))))).)).)))....---------.))))........ ( -16.60) >DroEre_CAF1 167843 98 + 1 AGCC-----GCUGAA----CAGAACAGCCUCCCAAACCACAGGCAUCGCAAAGUCUAACCGCAACAAUUGGCCAACAAGUUGUCGCUGGUCAAC---------AGUUCAUGCAACA ....-----((((((----(......((((..........))))........((...(((((.((((((........)))))).)).)))..))---------.))))).)).... ( -22.70) >DroYak_CAF1 165010 98 + 1 GAUC-----CGUCAA----CAGAAAAGCCUGCAAAACCACAGGCAUCGCAAAGUCUAACCGCAACAAUUGGCCAACAAGUUGUCGCUGGUCAAC---------AGUUCAUACAACA ....-----.(((((----.......(((((........)))))...((...........)).....)))))......(((((.((((.....)---------)))....))))). ( -20.50) >DroAna_CAF1 156746 102 + 1 UACCGACAUCCUCAAUCCUCUGAAAA-----AGAAACUUCAGGCAUCUCAAAGUCUAACCACAACAAUUGGCCAACAAGUUGUCGCUGGCCAAC---------UGCUCUGACAACA ..................(((.....-----)))....(((((((.......((......)).....(((((((.(........).))))))).---------))).))))..... ( -17.10) >consensus UACC_____CCUCAA____CAGAACAGCCUCCAAAACCACAGGCAUCGCAAAGUCUAACCGCAACAAUUGGCCAACAAGUUGUCGCUGGUCAAC_________AGUUCAUACAACA .....................(((................((((........))))...........(((((((.(........).)))))))............)))........ (-10.27 = -10.05 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:49 2006