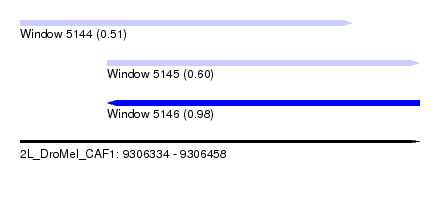
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,306,334 – 9,306,458 |
| Length | 124 |
| Max. P | 0.977741 |

| Location | 9,306,334 – 9,306,437 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
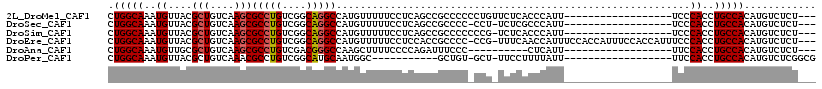
>2L_DroMel_CAF1 9306334 103 + 22407834 UCUGGCCGC--UGCAUUGUUGCCGCUUUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCCCCUGUUCUCACCCAUU-- ...(((.((--((.....((((((......))))))(((....(((....)))(((((....)))))))).........)))).)))..................-- ( -28.90) >DroSec_CAF1 135798 101 + 1 UCUGGCCGC--UGCAUUGUUGCCGCUUUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCC-CCU-UCUCGCCCAUU-- ...(((.((--((.....((((((......))))))(((....(((....)))(((((....)))))))).........)))).)))..-...-...........-- ( -28.90) >DroSim_CAF1 138896 102 + 1 UCUGGCCGC--UGCAUUGUUGCCGCUUUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCCCCCG-UCUCACCCAUU-- ...(((.((--((.....((((((......))))))(((....(((....)))(((((....)))))))).........)))).)))......-...........-- ( -28.90) >DroEre_CAF1 148021 103 + 1 UCUGGGCGC--UGCAUUGUUGCCGCUCUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCCACCGCCCC-CCG-UUUCAACCAUUUC ...(((((.--((.....((((((......))))))(((....(((....)))(((((....))))))))..........)).))))).-...-............. ( -28.60) >DroYak_CAF1 144285 90 + 1 UCUGGCCGC--UGCAUUGUUGCCGCUUUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUCCCU---------------UUUCACCCAUUUC ..(((((((--((((...((((((......))))))......((((....))))..)).)))).)))))..........---------------............. ( -26.10) >DroAna_CAF1 136505 95 + 1 UCUGGCCGCAUUGCAUUGUUGCCGCUUUUCUGGCAAAUGUUGCGCUGUCAAGCGCCUGUCGACGGGCCAAGCUUUUCCCCAGAUUUCCC----------CUCAUU-- (((((..(((.((((...((((((......))))))....)))).))).(((((((((....)))))...))))....)))))......----------......-- ( -29.00) >consensus UCUGGCCGC__UGCAUUGUUGCCGCUUUUCUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCC_CCG_UCUCACCCAUU__ ..((((.((...(((((..(((((......))))))))))...)).)))).(.(((((....))))))....................................... (-23.47 = -23.50 + 0.03)

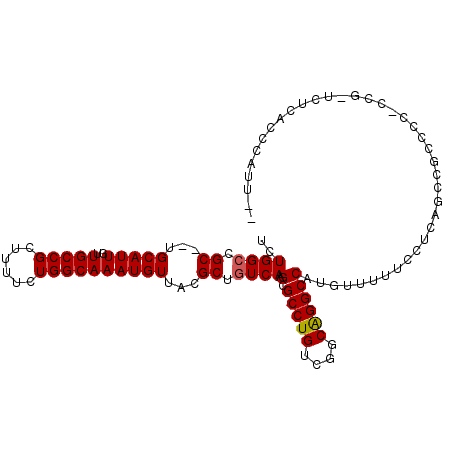
| Location | 9,306,361 – 9,306,458 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -16.87 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9306361 97 + 22407834 CUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCCCCUGUUCUCACCCAUU------------------UCCCACCUGCCACAUGUCUCU--- ..((((.......((((....))))..(((.((((((..(((........(((........)))........))).------------------.....)))))))))))))...--- ( -20.09) >DroSec_CAF1 135825 95 + 1 CUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCC-CCU-UCUCGCCCAUU------------------UCCCACCUGCCACAUGUCUCU--- .(((((..((...((((....))))......(((.(((..((........))))))))..-...-...........------------------...))..))))).........--- ( -19.40) >DroSim_CAF1 138923 96 + 1 CUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCCCCCG-UCUCACCCAUU------------------UCCCACCUGCCACAUGUCUCU--- .(((((..((...((((....))))...(.(((..(((..((........))...)))...)))-.).........------------------...))..))))).........--- ( -19.70) >DroEre_CAF1 148048 113 + 1 CUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCCACCGCCCC-CCG-UUUCAACCAUUUCCACCAUUUCCACCAUUUCCCACCUGCCACAUGUCUCU--- .(((((..((....(((....)))(((((....)))))......................-...-................................))..))))).........--- ( -18.90) >DroAna_CAF1 136534 87 + 1 CUGGCAAAUGUUGCGCUGUCAAGCGCCUGUCGACGGGCCAAGCUUUUCCCCAGAUUUCCC----------CUCAUU------------------UUCCACCUGCCACAUGUCUCU--- .(((((..((..(((((....)))))(((..((..(((...)))..))..))).......----------......------------------...))..))))).........--- ( -21.70) >DroPer_CAF1 161155 87 + 1 CUGGCAAAUGUUACGCUGUCAAACGCCUGUCGGCAUGCAAUGGC-----------GCUGU-GCU-UUCCUUUUAUU------------------UUCCACCUGCCACAUGUCUCGGCG .(((((..........)))))..((((....((((((...((((-----------(..((-(..-...........------------------...))).)))))))))))..)))) ( -19.99) >consensus CUGGCAAAUGUUACGCUGUCAAGCGCCUGUCGGCAGGCCAUGUUUUUCCUCAGCCGCCCC_CCG_UCUCACCCAUU__________________UCCCACCUGCCACAUGUCUCU___ .(((((..((....(((....)))(((((....)))))...........................................................))..)))))............ (-16.87 = -17.07 + 0.20)
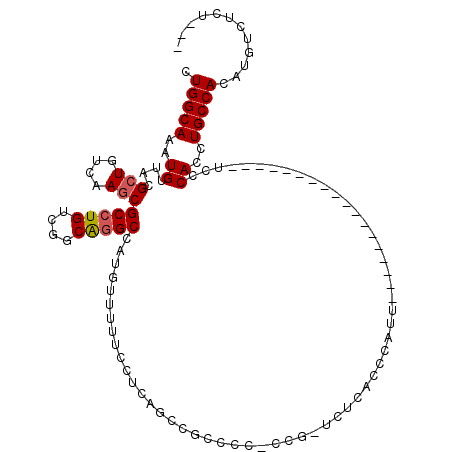
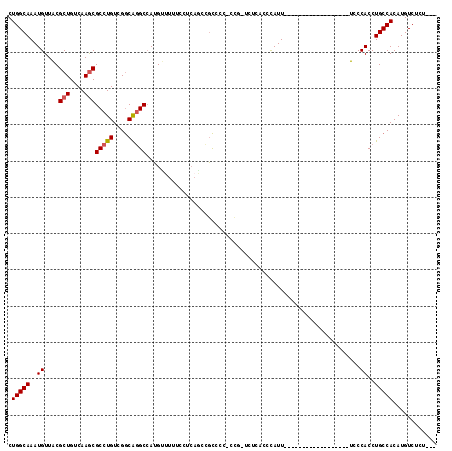
| Location | 9,306,361 – 9,306,458 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9306361 97 - 22407834 ---AGAGACAUGUGGCAGGUGGGA------------------AAUGGGUGAGAACAGGGGGGCGGCUGAGGAAAAACAUGGCCUGCCGACAGGCGCUUGACAGCGUAACAUUUGCCAG ---.........(((((((((...------------------...............(..((((((((.(......).))))).)))..)..(((((....)))))..))))))))). ( -31.00) >DroSec_CAF1 135825 95 - 1 ---AGAGACAUGUGGCAGGUGGGA------------------AAUGGGCGAGA-AGG-GGGGCGGCUGAGGAAAAACAUGGCCUGCCGACAGGCGCUUGACAGCGUAACAUUUGCCAG ---.........(((((((((..(------------------..((..((((.-..(-..((((((((.(......).))))).)))..).....)))).))...)..))))))))). ( -31.40) >DroSim_CAF1 138923 96 - 1 ---AGAGACAUGUGGCAGGUGGGA------------------AAUGGGUGAGA-CGGGGGGGCGGCUGAGGAAAAACAUGGCCUGCCGACAGGCGCUUGACAGCGUAACAUUUGCCAG ---.........(((((((((...------------------...........-...(..((((((((.(......).))))).)))..)..(((((....)))))..))))))))). ( -31.00) >DroEre_CAF1 148048 113 - 1 ---AGAGACAUGUGGCAGGUGGGAAAUGGUGGAAAUGGUGGAAAUGGUUGAAA-CGG-GGGGCGGUGGAGGAAAAACAUGGCCUGCCGACAGGCGCUUGACAGCGUAACAUUUGCCAG ---.........(((((((((.....((((...(((.((....)).)))...)-)((-.((((.(((.........))).)))).))..)).(((((....)))))..))))))))). ( -38.30) >DroAna_CAF1 136534 87 - 1 ---AGAGACAUGUGGCAGGUGGAA------------------AAUGAG----------GGGAAAUCUGGGGAAAAGCUUGGCCCGUCGACAGGCGCUUGACAGCGCAACAUUUGCCAG ---.........(((((((((...------------------..(((.----------(((....(..((......))..)))).)))....(((((....)))))..))))))))). ( -30.70) >DroPer_CAF1 161155 87 - 1 CGCCGAGACAUGUGGCAGGUGGAA------------------AAUAAAAGGAA-AGC-ACAGC-----------GCCAUUGCAUGCCGACAGGCGUUUGACAGCGUAACAUUUGCCAG ............(((((((((...------------------.......(...-...-.).((-----------((..(((.(((((....))))).)))..))))..))))))))). ( -24.10) >consensus ___AGAGACAUGUGGCAGGUGGGA__________________AAUGGGUGAGA_CGG_GGGGCGGCUGAGGAAAAACAUGGCCUGCCGACAGGCGCUUGACAGCGUAACAUUUGCCAG ............(((((((((.......................................(((((((..(......)..)))).))).....(((((....)))))..))))))))). (-22.83 = -23.83 + 1.00)

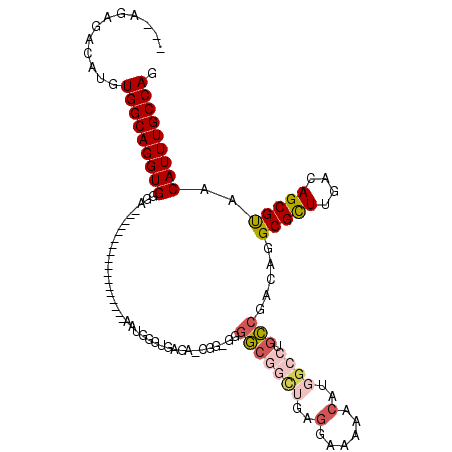
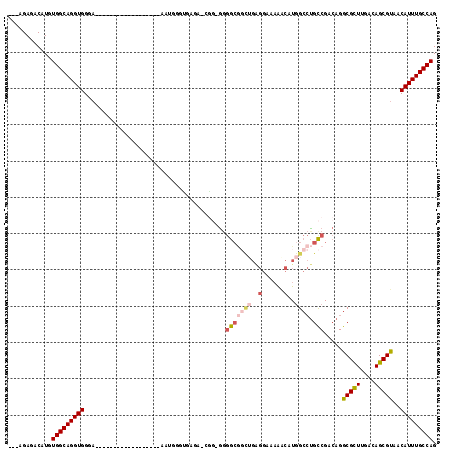
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:44 2006