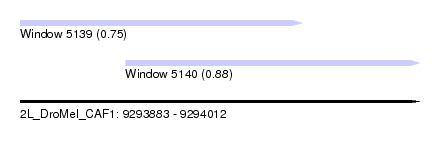
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,293,883 – 9,294,012 |
| Length | 129 |
| Max. P | 0.882763 |

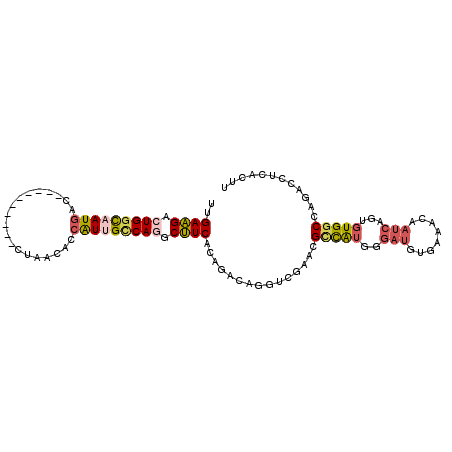
| Location | 9,293,883 – 9,293,974 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9293883 91 + 22407834 ---AAUGAAAUGGUUUAGUGAAGUAUAAUCA-AAAGGCUUGAAGACUGGUAAUGAC----------CUGACACCAUUGCCAGGCUUCACAGACCGGUCGAAUGCC ---..(((..((((((.(((((((....(((-(.....))))...(((((((((..----------.......)))))))))))))))))))))).)))...... ( -29.40) >DroSec_CAF1 123622 91 + 1 ---AAUGAAAUGGUUUAGUAAAGUAUAAUCA-AAAGGCUUGAAGACUGGCAAUGAC----------CUAACACCAUUGUCAUGCUUCACAGACCGGUCGAAUGCC ---..(((..((((((.((.((((((..(((-(.....)))).....(((((((..----------.......))))))))))))).)))))))).)))...... ( -20.20) >DroSim_CAF1 126331 91 + 1 ---AAUGAAAUGGUUUAGUGAAGUAUAAUCA-AAAGGCUUGAAGACUGGCAAUGAC----------CUAACACAAUUGCCAUGCUUCACAGAUCGGUCGAAUGUC ---..(((..((((((.(((((((((..(((-(.....)))).....((((((...----------........))))))))))))))))))))).)))...... ( -24.10) >DroEre_CAF1 133767 91 + 1 ---AAUGAAAUGAUUUAGUGAAGUAUAAUCA-AAAGGCUUGAAGGCUGGCCAUGAC----------CUAACAGCAUUGCCAGGCUUCACAGACAGGUGGAACGCC ---..............(((((((....(((-(.....))))...(((((.(((.(----------......)))).)))))))))))).....((((...)))) ( -22.40) >DroYak_CAF1 131535 91 + 1 ---AAUGAAAUGAUUUAGUGAAGUAUAAUCA-AAAGGCUUGAAGGCUGGCCAUGAC----------CUAGCAGCGUUGCCAGACUUCACAGACAGGUGGAACGCC ---..............(((((((....(((-(.....))))...(((((.(((.(----------......)))).)))))))))))).....((((...)))) ( -21.50) >DroPer_CAF1 146463 97 + 1 GACGAUGAAAU-----AACG--AUAUAAUCAAAAAGGCCCGAGGGCUGUCAAUGACUCGCUGCAAUCCAGCGACAUUGCCAGACCUCACAGACAGGA-GACCGUU ((((.......-----....--..................((((.(((.(((((..((((((.....))))))))))).))).)))).......(..-..))))) ( -26.60) >consensus ___AAUGAAAUGGUUUAGUGAAGUAUAAUCA_AAAGGCUUGAAGACUGGCAAUGAC__________CUAACACCAUUGCCAGGCUUCACAGACAGGUCGAACGCC ...................................(((..((((.(((((((((...................))))))))).)))).......(......)))) (-14.60 = -14.33 + -0.27)

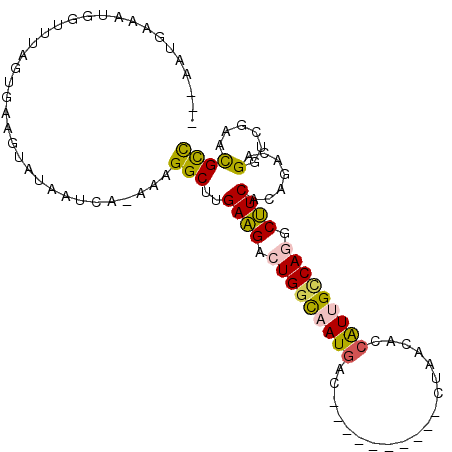
| Location | 9,293,917 – 9,294,012 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -18.67 |
| Energy contribution | -19.39 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9293917 95 + 22407834 UUGAAGACUGGUAAUGAC----------CUGACACCAUUGCCAGGCUUCACAGACCGGUCGAAUGCCAUUGGAUGUGAAACAAUCAGUGUGGCCAGACCUCACUU .(((((.(((((((((..----------.......))))))))).)))))......((((....(((((..(((........)))...)))))..))))...... ( -33.30) >DroSec_CAF1 123656 95 + 1 UUGAAGACUGGCAAUGAC----------CUAACACCAUUGUCAUGCUUCACAGACCGGUCGAAUGCCAUUGGAUGUGAAACAAUCAGUGUGGCCAGACCUCACUU .(((((..((((((((..----------.......))))))))..)))))......((((....(((((..(((........)))...)))))..))))...... ( -29.50) >DroSim_CAF1 126365 95 + 1 UUGAAGACUGGCAAUGAC----------CUAACACAAUUGCCAUGCUUCACAGAUCGGUCGAAUGUCAUUGGAUGUGAAACAAUCAGUGUGGCCAGACCUCACUU .(((((..(((((((...----------........)))))))..)))))......((((....(((((..(((........)))...)))))..))))...... ( -27.10) >DroEre_CAF1 133801 95 + 1 UUGAAGGCUGGCCAUGAC----------CUAACAGCAUUGCCAGGCUUCACAGACAGGUGGAACGCCAUGGGAUGUGAAACAAUCAGUGUGCCCAGACCUCACUU .(((.((((((((((((.----------......(((((.(((((((((((......)))))..))).)))))))).......))).)).).)))).)))))... ( -28.14) >DroYak_CAF1 131569 95 + 1 UUGAAGGCUGGCCAUGAC----------CUAGCAGCGUUGCCAGACUUCACAGACAGGUGGAACGCCAUGGGAUGAGAAACAAUCAGUGUGGCCAGAGCCCACUU .....((((((((((..(----------(((...(((((.(((..((........)).))))))))..)))).(((.......)))..))))))..))))..... ( -30.10) >DroPer_CAF1 146494 98 + 1 CCGAGGGCUGUCAAUGACUCGCUGCAAUCCAGCGACAUUGCCAGACCUCACAGACAGGA-GACCGUUGUGGCAUAGGGAACAU---GUCUGGCAUCAGCG---CU ...((.((((..((((..((((((.....))))))))))(((((((.((((((.(.(..-..).)))))))....(....)..---)))))))..)))).---)) ( -36.50) >consensus UUGAAGACUGGCAAUGAC__________CUAACACCAUUGCCAGGCUUCACAGACAGGUCGAACGCCAUGGGAUGUGAAACAAUCAGUGUGGCCAGACCUCACUU ..((((.(((((((((...................))))))))).))))...............(((((..(((........)))...)))))............ (-18.67 = -19.39 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:39 2006