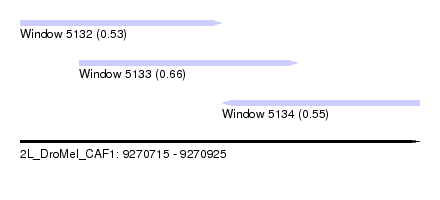
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,270,715 – 9,270,925 |
| Length | 210 |
| Max. P | 0.662283 |

| Location | 9,270,715 – 9,270,821 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9270715 106 + 22407834 GAGCAACUUUUCCUCGAAGCGGCAGAAGCAGAUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGUUUGGUCCUUUGGCGGAAAGAA ......((((((.((((((.(((.((((.(..((((((....))))))..).)))).)).(((.(((.(((.......)))..))).)))))))))).)))))).. ( -29.00) >DroSec_CAF1 100636 106 + 1 GAGCAACUUUUCCUCGAAGCGGCAAAAGCAGAUUGGGUUAAAGCCUAAAAUGCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAA ......((((((.((((((.(((.((((((..((((((....))))))..))))))..((((((....(((.......))).))))))))))))))).)))))).. ( -38.40) >DroSim_CAF1 102737 106 + 1 GAGCAACUUUUCCUCGAAGCGGCAAAAGCAGAUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAA ......((((((.((((((.(((.((((.(..((((((....))))))..).))))..((((((....(((.......))).))))))))))))))).)))))).. ( -31.80) >DroEre_CAF1 106736 106 + 1 GUGCAACCUUUUCUCGCAGCGGCAGUAUCAGAUUGGGUUAAAGCCCAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAUGUUGGUCCUUUGGCGGAAAGAA ...(((((((((((.(((((..(((.......((((((....))))))((....))......)))...))))))))))))....)))).(((......)))..... ( -26.90) >DroYak_CAF1 107923 106 + 1 GAGCAACUUUUCCUCGCAGCGGCAGUAGCAGAUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGUUUGGUCCUUUGGCGGGUAGAA ........(((.(((((.((.((....))...((((((....)))))).........)).(((.(((.(((.......)))..))).)))......))))).))). ( -24.60) >consensus GAGCAACUUUUCCUCGAAGCGGCAGAAGCAGAUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAA .......((((((((((((.(((.........((((((....))))))..........((((((....(((.......))).))))))))))))))).)))))).. (-25.32 = -25.88 + 0.56)

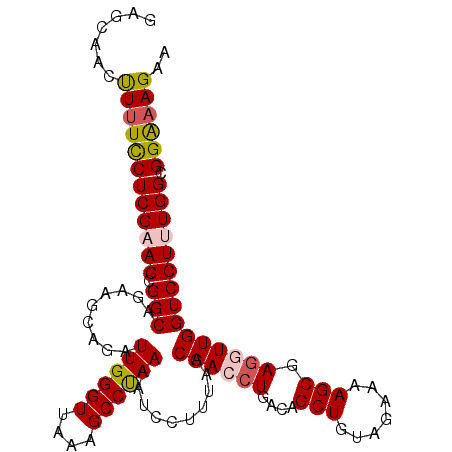
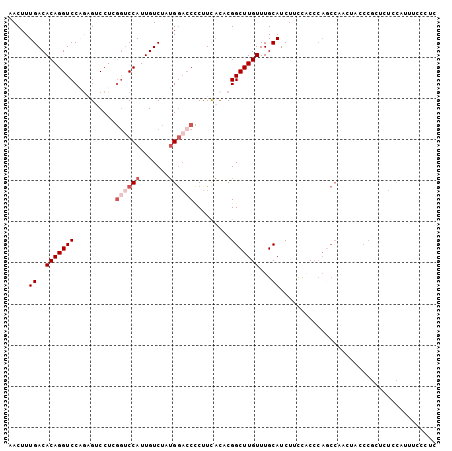
| Location | 9,270,746 – 9,270,861 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9270746 115 + 22407834 AUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGUUUGGUCCUUUGGCGGAAAGAAGAGGGAAAUGAAGAGCGGGUAGUUGGUUGGGUGGAAGAUG .((((((....)))))).((((.(((((.(((((.(.(((.......)))........((((((..(.....)..))))))........)))))).)))))..))))........ ( -29.50) >DroSec_CAF1 100667 115 + 1 AUUGGGUUAAAGCCUAAAAUGCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAAGAGGGAAAUGGAGAGCGGGUAGUUGGCUGGGUGAAAGAUG (((.((((((.((((.....(((((..((((((....(((.......))).)))))).((((((..(.....)..)))))).....)))))))))..)))))).)))........ ( -30.50) >DroSim_CAF1 102768 115 + 1 AUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAAGAGGGAAAUGGAGAGCGGGUAGUUGGCUGGGUGAAAGAUG (((.((((((.((((....(((.....((((((....(((.......))).)))))).((((((..(.....)..))))))...)))....))))..)))))).)))........ ( -31.30) >DroEre_CAF1 106767 115 + 1 AUUGGGUUAAAGCCCAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAUGUUGGUCCUUUGGCGGAAAGAAGAAGGAAAUGGAGAACGGGCAGUGGGCUGGGCAGGAGAUG .((((((....))))))..(((((((((.(((..((((((.......)))..)))..)((((((..(.....)..))))))...............))..)))))..)))).... ( -30.50) >DroYak_CAF1 107954 104 + 1 AUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGUUUGGUCCUUUGGCGGGUAGAAGAAGGAAGUGGAGAAUGG-----------GGCAGUAGAUG ...........((((....(((........((.(((.(((.......)))..))).))((((((..(.....)..))))))...))).....)-----------)))........ ( -25.10) >consensus AUUGGGUUAAAGCCUAAAAUCCUUUAGCAACCUGACAGCUGUAGAAAAGCGAGGUUGGUCCUUUGGCGGAAAGAAGAGGGAAAUGGAGAGCGGGUAGUUGGCUGGGUGGAAGAUG .((((((....))))))....(((((.((((((....(((.......))).)))))).((((((..(.....)..))))))..)))))........................... (-22.30 = -22.34 + 0.04)

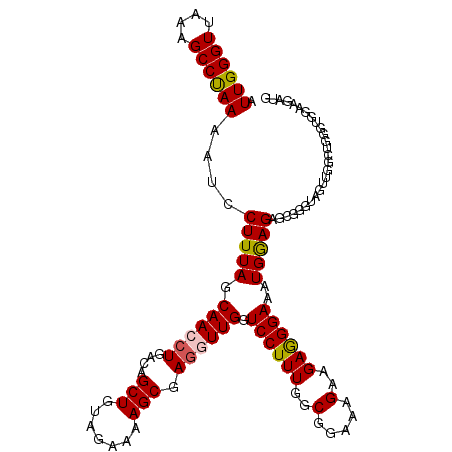
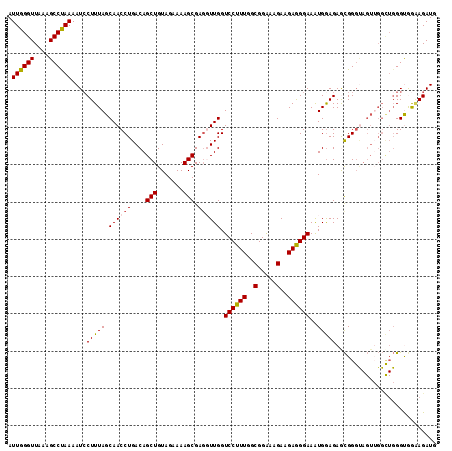
| Location | 9,270,821 – 9,270,925 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -9.29 |
| Energy contribution | -10.46 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
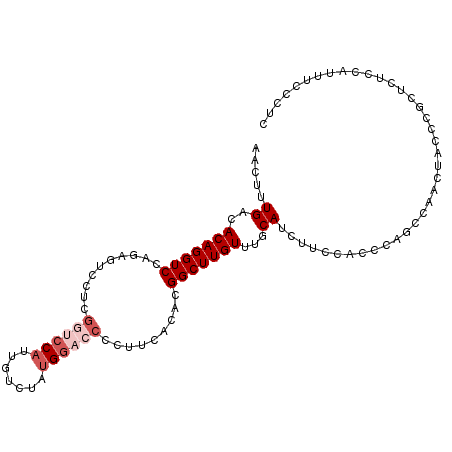
>2L_DroMel_CAF1 9270821 104 - 22407834 AACUUUGACACAGGUCCAAAGUCCUCGGUCCAUUGUCUAUGGACCCCUUCACACGGCUUGUUUGCAUCUUCCACCCAACCAACUACCCGCUCUUCAUUUCCCUC .((((((((....)).))))))....(((((((.....))))))).........((.(((..((.......))..)))))........................ ( -18.40) >DroSec_CAF1 100742 104 - 1 AACUUUGACACAGGUCCAGAGUCCUCGGUCCAUUGUCUAUGGACCCCUUCACACGGCUUGUUUGCAUCUUUCACCCAGCCAACUACCCGCUCUCCAUUUCCCUC ............((...(((((....(((((((.....))))))).........((((.(..((.......))..)))))........)))))))......... ( -21.80) >DroSim_CAF1 102843 104 - 1 AACUUUGACACAGGUCCAGAGUCCUCGGUCCAUUGUCUAUGGACCCCUUCACACGGCUUGUUUGCAUCUUUCACCCAGCCAACUACCCGCUCUCCAUUUCCCUC ............((...(((((....(((((((.....))))))).........((((.(..((.......))..)))))........)))))))......... ( -21.80) >DroEre_CAF1 106842 104 - 1 CACUUUGACACAGGUCCAGCGACCUGGCGCCAUUGUCUCUGGACCCCUUCACACGGCUUGUUUGCAUCUCCUGCCCAGCCCACUGCCCGUUCUCCAUUUCCUUC ......(((...(((((((.(((.((....))..))).))))))).........((((.(...(((.....)))).))))........)))............. ( -23.90) >DroYak_CAF1 108029 93 - 1 AACUUUGACACAGGUCCCGAGUCCUGGGUCCAUUGUCUCUGAAACUCCUCACACGGCUUGUUUGCAUCUACUGCC-----------CCAUUCUCCACUUCCUUC ......((((..((.((((.....)))).))..)))).................(((..((........)).)))-----------.................. ( -16.50) >DroAna_CAF1 101013 84 - 1 AACUUUGACACAGGUCCUGUGUCCUGUGUCCAUUGUCCCCGGUCCACUUUCGCCGGCUUGUUUGCAUCUCUUCCGCAGCCGAGU-------------------- ......((((((((........))))))))........(((((........)))))((((.((((.........)))).)))).-------------------- ( -25.50) >consensus AACUUUGACACAGGUCCAGAGUCCUCGGUCCAUUGUCUAUGGACCCCUUCACACGGCUUGUUUGCAUCUUCCACCCAGCCAACUACCCGCUCUCCAUUUCCCUC .....((..(((((((..........((((((.......)))))).........)))))))...))...................................... ( -9.29 = -10.46 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:33 2006