
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,245,579 – 9,245,681 |
| Length | 102 |
| Max. P | 0.985456 |

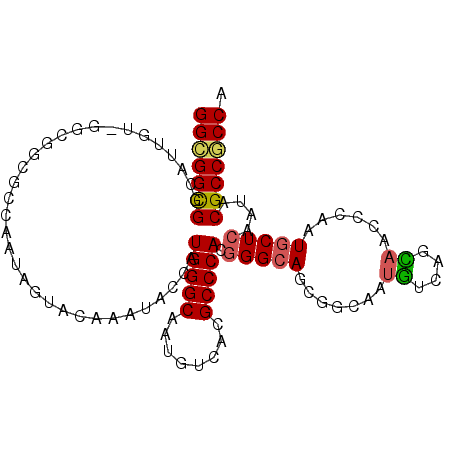
| Location | 9,245,579 – 9,245,681 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.39 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
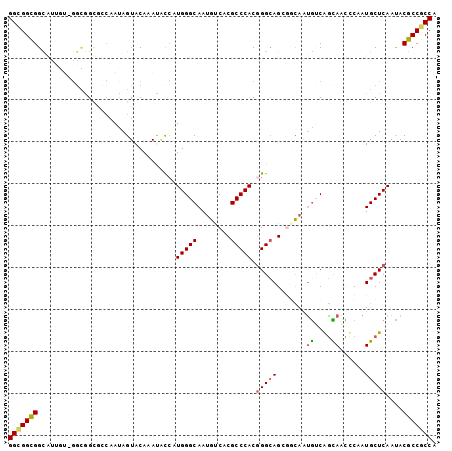
>2L_DroMel_CAF1 9245579 102 + 22407834 GGCGGUGGCAUUGUGGGCGGAGUCAACAGUACAAACACAAUGGGCAAUGUCACGCCCACGGGCAGCGGCAAUGUCAGCAACUCCAUGCUCAACACGCCACCG ..(((((((....((((((((((.....((....))......((((.((((.((((....))).).)))).))))....)))))..)))))....))))))) ( -40.80) >DroPse_CAF1 92574 99 + 1 GGCGGCGGCAUUGUCGGUGGCCCCAAUAGUACAAAUGCCAUGGGCAAUGUCACGCCCACGGGCAG---UAAUGUCGGCAACCCUAUGCUCAAUACGCCGCCG (((((((((((((((.(((((...............))))).)))))))))..(((....))).(---((.((..((((......)))))).))))))))). ( -46.76) >DroWil_CAF1 135838 96 + 1 GGUGGCGGC------GGCAGCGGAAAUAGUACAAAUAUCAUGGGCAAUGCCACGCCCACGGGAAGCGGCAAUGGCAAUAAUGCAAUGCUCAAUACGCCACCA (((((((((------....))...................((((((.((((.(.((....))..).))))...(((....)))..))))))...))))))). ( -35.70) >DroYak_CAF1 81692 102 + 1 GGCGGCGGUAUCGUGGGCGGGGUCAACAGUACAAACACCAUGGGCAACGUCACGCCCACGGGCAGCGGCAAUGUCAGCAACUCCAUGCUCAACACGCCGCCA (((((((((...(((((((.(((.............)))...(....)....)))))))(((((..((...((....))...)).))))).)).))))))). ( -43.92) >DroMoj_CAF1 134894 93 + 1 GGCGGCGGU---------AGCGGCAAUAGUACAAAUUCCAUGGGCAAUGCCACGCCCACCGGCAGCGGCAGCAGCGGUAAUCCAAUGCUCAAUACGCCGCCA (((((((..---------...((.(((.......))))).((((((.((((.((((....))).).)))).....((....))..))))))...))))))). ( -37.00) >DroPer_CAF1 92698 99 + 1 GGCGGCGGCAUUGUCGGUGGCCCCAAUAGUACAAAUGCCAUGGGCAAUGUCACGCCCACGGGCAG---UAAUGUCGGCAACCCUAUGCUCAAUACGCCGCCG (((((((((((((((.(((((...............))))).)))))))))..(((....))).(---((.((..((((......)))))).))))))))). ( -46.76) >consensus GGCGGCGGCAUUGU_GGCGGCGCCAAUAGUACAAAUACCAUGGGCAAUGUCACGCCCACGGGCAGCGGCAAUGUCAGCAACCCAAUGCUCAAUACGCCGCCA (((((((.................................(((((........))))).(((((.......((....))......)))))....))))))). (-27.44 = -27.39 + -0.05)

| Location | 9,245,579 – 9,245,681 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
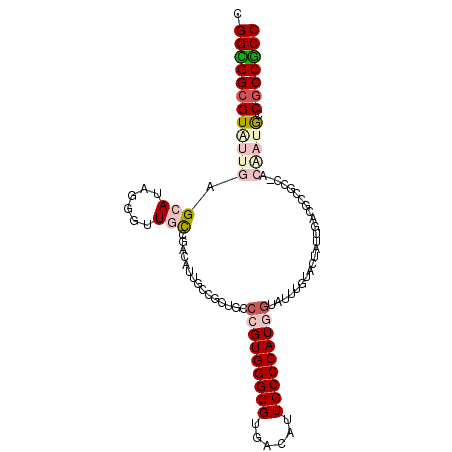
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -27.71 |
| Energy contribution | -29.13 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
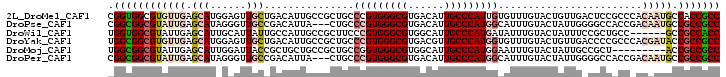
>2L_DroMel_CAF1 9245579 102 - 22407834 CGGUGGCGUGUUGAGCAUGGAGUUGCUGACAUUGCCGCUGCCCGUGGGCGUGACAUUGCCCAUUGUGUUUGUACUGUUGACUCCGCCCACAAUGCCACCGCC (((((((((..((.((..((((((...((((.(((....(((.(((((((......))))))).).))..))).)))))))))))).))..))))))))).. ( -42.90) >DroPse_CAF1 92574 99 - 1 CGGCGGCGUAUUGAGCAUAGGGUUGCCGACAUUA---CUGCCCGUGGGCGUGACAUUGCCCAUGGCAUUUGUACUAUUGGGGCCACCGACAAUGCCGCCGCC .((((((((((((.......((((.((((.(.((---(((((.(((((((......)))))))))))...))).).)))))))).....))))).))))))) ( -43.10) >DroWil_CAF1 135838 96 - 1 UGGUGGCGUAUUGAGCAUUGCAUUAUUGCCAUUGCCGCUUCCCGUGGGCGUGGCAUUGCCCAUGAUAUUUGUACUAUUUCCGCUGCC------GCCGCCACC ((((((((....(.(((..(((....)))...))))......((((((((......))))))))......................)------))))))).. ( -33.50) >DroYak_CAF1 81692 102 - 1 UGGCGGCGUGUUGAGCAUGGAGUUGCUGACAUUGCCGCUGCCCGUGGGCGUGACGUUGCCCAUGGUGUUUGUACUGUUGACCCCGCCCACGAUACCGCCGCC .((((((((((((.(..(((.(((...((((.(((....(((((((((((......))))))))).))..))).))))))).)))..).))))).))))))) ( -45.10) >DroMoj_CAF1 134894 93 - 1 UGGCGGCGUAUUGAGCAUUGGAUUACCGCUGCUGCCGCUGCCGGUGGGCGUGGCAUUGCCCAUGGAAUUUGUACUAUUGCCGCU---------ACCGCCGCC .(((((((.....((((.(((....))).))))..)))))))(((((..((((((..(..((.......))..)...)))))).---------.)))))... ( -39.90) >DroPer_CAF1 92698 99 - 1 CGGCGGCGUAUUGAGCAUAGGGUUGCCGACAUUA---CUGCCCGUGGGCGUGACAUUGCCCAUGGCAUUUGUACUAUUGGGGCCACCGACAAUGCCGCCGCC .((((((((((((.......((((.((((.(.((---(((((.(((((((......)))))))))))...))).).)))))))).....))))).))))))) ( -43.10) >consensus CGGCGGCGUAUUGAGCAUAGGGUUGCCGACAUUGCCGCUGCCCGUGGGCGUGACAUUGCCCAUGGUAUUUGUACUAUUGACGCCGCC_ACAAUGCCGCCGCC .((((((((((((.(((......)))...............(((((((((......)))))))))........................))))).))))))) (-27.71 = -29.13 + 1.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:26 2006