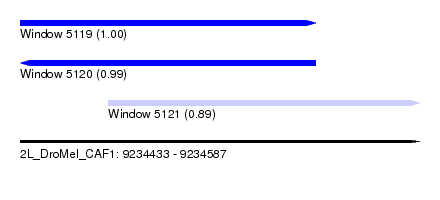
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,234,433 – 9,234,587 |
| Length | 154 |
| Max. P | 0.995820 |

| Location | 9,234,433 – 9,234,547 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
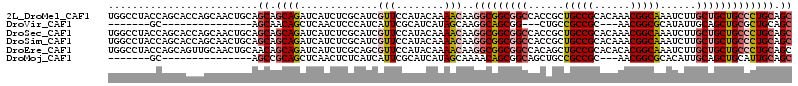
>2L_DroMel_CAF1 9234433 114 + 22407834 UGGCCUACCAGCACCAGCAACUGCAGCAGCAGAUCAUCUCGCAUCGUUCCAUACAAAACAAGGCGGCGGCCACCGCUGCCGCACAAACGGCAAAUCUUGCUGCUGCCCUGCAGC (((....)))......(((...((((((((((((....(((....(((........)))...((((((((....)))))))).....)))...))).)))))))))..)))... ( -40.30) >DroVir_CAF1 106456 86 + 1 -------GC---------------AGCAACAGCUCAACUCCCAUCAUUCGCAUCAUAGCAAGGCAGCGG---CUGCCGCCGC---AACGGCGCAUAUUGCAGCUGCGCUGCAGC -------((---------------(((......................((......))......((((---(((((((((.---..)))))......)))))))))))))... ( -32.80) >DroSec_CAF1 64259 114 + 1 UGGCCUACCAGCACCAGCAACUGCAGCAGCAGAUCAUCUCGCAUCGUUCCAUACAAAACAAGGCGGCGGCCACCGCUGCCGCACAAACGGCAAAUCUUGCUGCUGCCCUGCAGC (((....)))......(((...((((((((((((....(((....(((........)))...((((((((....)))))))).....)))...))).)))))))))..)))... ( -40.30) >DroSim_CAF1 66126 114 + 1 UGGCCUACCAGCACCAGCAACUGCAGCAGCAGAUCAUCUCGCAUCGUUCCAUACAAAACAAGGCGGCGGCCACCGCUGCCGCACAAACGGCAAAUCUUGCUGCUGCCCUGCAGC (((....)))......(((...((((((((((((....(((....(((........)))...((((((((....)))))))).....)))...))).)))))))))..)))... ( -40.30) >DroEre_CAF1 71092 114 + 1 UGGCCUACCAGCAGUUGCAACUGCAACAGCAGAUCAUCUCGCAGCGUUCCAUACAAAACAAGGCGGCGGCCACAGCUGCCGCACACACGGCAAAUCUUGCUGCUGCCCUGCAGC (((....)))...((((((.((((....))))........(((((.................((((((((....))))))))......((((.....)))))))))..)))))) ( -41.50) >DroMoj_CAF1 117028 89 + 1 -------GC---------------AGCCGCAGCUCAACUCUCAUCAUUCGCAUCAUAGCAAAACAGCGGCAGCUGCCGCCGC---AACGGCGCACAUUGCAGCUGCAUUGCAGC -------((---------------((..(((((((((............((......))......(((((....)))((((.---..))))))...))).)))))).))))... ( -30.20) >consensus UGGCCUACCAGCACCAGCAACUGCAGCAGCAGAUCAUCUCGCAUCGUUCCAUACAAAACAAGGCGGCGGCCACCGCUGCCGCACAAACGGCAAAUCUUGCUGCUGCCCUGCAGC .........................((.((((.............(((........)))..(((((((((......(((((......)))))......))))))))))))).)) (-24.04 = -24.68 + 0.64)
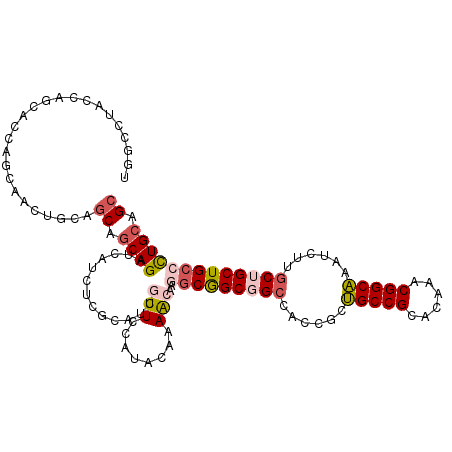

| Location | 9,234,433 – 9,234,547 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -46.57 |
| Consensus MFE | -31.57 |
| Energy contribution | -31.30 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9234433 114 - 22407834 GCUGCAGGGCAGCAGCAAGAUUUGCCGUUUGUGCGGCAGCGGUGGCCGCCGCCUUGUUUUGUAUGGAACGAUGCGAGAUGAUCUGCUGCUGCAGUUGCUGGUGCUGGUAGGCCA ((.((((.(((((((((.((((((((((....))))))(((((....)))))(((((.((((.....)))).)))))..)))))))))))))..))))....)).((....)). ( -51.80) >DroVir_CAF1 106456 86 - 1 GCUGCAGCGCAGCUGCAAUAUGCGCCGUU---GCGGCGGCAG---CCGCUGCCUUGCUAUGAUGCGAAUGAUGGGAGUUGAGCUGUUGCU---------------GC------- ...(((((((((((.((((...(.(((((---(((((((...---))))))).((((......))))..)))))).))))))))).))))---------------))------- ( -38.40) >DroSec_CAF1 64259 114 - 1 GCUGCAGGGCAGCAGCAAGAUUUGCCGUUUGUGCGGCAGCGGUGGCCGCCGCCUUGUUUUGUAUGGAACGAUGCGAGAUGAUCUGCUGCUGCAGUUGCUGGUGCUGGUAGGCCA ((.((((.(((((((((.((((((((((....))))))(((((....)))))(((((.((((.....)))).)))))..)))))))))))))..))))....)).((....)). ( -51.80) >DroSim_CAF1 66126 114 - 1 GCUGCAGGGCAGCAGCAAGAUUUGCCGUUUGUGCGGCAGCGGUGGCCGCCGCCUUGUUUUGUAUGGAACGAUGCGAGAUGAUCUGCUGCUGCAGUUGCUGGUGCUGGUAGGCCA ((.((((.(((((((((.((((((((((....))))))(((((....)))))(((((.((((.....)))).)))))..)))))))))))))..))))....)).((....)). ( -51.80) >DroEre_CAF1 71092 114 - 1 GCUGCAGGGCAGCAGCAAGAUUUGCCGUGUGUGCGGCAGCUGUGGCCGCCGCCUUGUUUUGUAUGGAACGCUGCGAGAUGAUCUGCUGUUGCAGUUGCAACUGCUGGUAGGCCA (((((......))))).....(((((((....)))))))...((((((((..(((((..(((.....)))..))))).......((.(((((....))))).)).))).))))) ( -45.30) >DroMoj_CAF1 117028 89 - 1 GCUGCAAUGCAGCUGCAAUGUGCGCCGUU---GCGGCGGCAGCUGCCGCUGUUUUGCUAUGAUGCGAAUGAUGAGAGUUGAGCUGCGGCU---------------GC------- (((((((((..((.((.....))))))))---))))).((((((((.(((..(((((......))))).(((....))).))).))))))---------------))------- ( -40.30) >consensus GCUGCAGGGCAGCAGCAAGAUUUGCCGUUUGUGCGGCAGCGGUGGCCGCCGCCUUGUUUUGUAUGGAACGAUGCGAGAUGAUCUGCUGCUGCAGUUGCUGGUGCUGGUAGGCCA ........(((((((((.....((((((....))))))(((((....)))))(((((..(((.....)))..)))))......)))))))))...................... (-31.57 = -31.30 + -0.27)

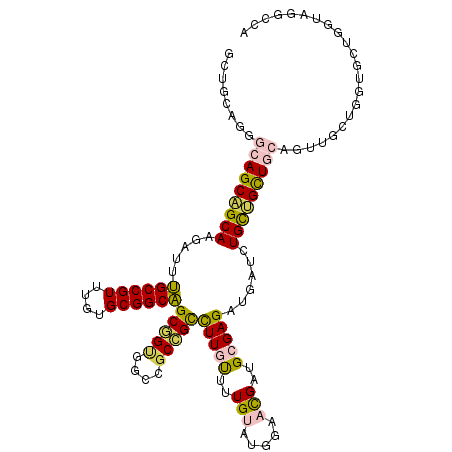

| Location | 9,234,467 – 9,234,587 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -42.39 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9234467 120 + 22407834 CAUCUCGCAUCGUUCCAUACAAAACAAGGCGGCGGCCACCGCUGCCGCACAAACGGCAAAUCUUGCUGCUGCCCUGCAGCGAUCGGCUGCCAUUAUGAAUGCGGUGGCAUCGCCGAUCUC ......((...((.....))....((.(((((((((......(((((......)))))......))))))))).))..))(((((((((((((..(....)..))))))..))))))).. ( -48.90) >DroVir_CAF1 106468 114 + 1 CAACUCCCAUCAUUCGCAUCAUAGCAAGGCAGCGG---CUGCCGCCGC---AACGGCGCAUAUUGCAGCUGCGCUGCAGCGCUCGGCCGCCAUUAUGAAUGCGGUGGCAUCGCCGAUUCC ........(((..(((((((((((...(((.((((---(....)))))---..((((((....((((((...)))))))))).)))))....))))).)))))).(((...))))))... ( -45.90) >DroPse_CAF1 80164 111 + 1 CAACUCUCAUCAUUCUCUCCA---CAAGGCAGCGGCCGC---CGCCGCAGCCACGGCCAACAUCGCAGCUGCGCUGCAACGCUCGG---CCAUUAUGAAUGCGGUGGCGUCGCCGAUCUC ................((((.---...)).))(((((((---(((((((..((.((((......(((((...))))).......))---))....))..))))))))))..))))..... ( -41.72) >DroMoj_CAF1 117040 114 + 1 CAACUCUCAUCAUUCGCAUCAUAGCAAAACAGCGGCAGCUGCCGCCGC---AACGGCGCACAUUGCAGCUGCAUUGCAGCGAUCGG---CCAUUAUGAACGCGGUGGCAUCGCCGAUUCC ........(((....(((.....((......)).(((((((((((((.---..)))))......))))))))..))).(((((..(---((((..(....)..)))))))))).)))... ( -40.20) >DroAna_CAF1 64618 117 + 1 CAACUCGCACCAUUCUAUCCAAAAUAAGGCAGCGGCCAC---UGCCGCUCACCCGACAAACAUUGCAGCCGCGCUGCAGCGAUCGACCGCCAUUAUGAAUGCGGUGGCAUCGCCGAUCUC ......((...................((.((((((...---.))))))...)).........((((((...))))))))((((.(((((..........)))))(((...))))))).. ( -35.90) >DroPer_CAF1 80252 111 + 1 CAACUCUCAUCAUUCUCUCCA---CAAGGCAGCGGCCGC---CGCCGCAGCCACGGCCAACAUCGCAGCUGCGCUGCAACGCUCGG---CCAUUAUGAAUGCGGUGGCGUCGCCGAUCUC ................((((.---...)).))(((((((---(((((((..((.((((......(((((...))))).......))---))....))..))))))))))..))))..... ( -41.72) >consensus CAACUCUCAUCAUUCUCUCCA_A_CAAGGCAGCGGCCGC___CGCCGCA__AACGGCAAACAUUGCAGCUGCGCUGCAGCGAUCGG___CCAUUAUGAAUGCGGUGGCAUCGCCGAUCUC ...............................(((((.......))))).....((((...((((((.(((((...)))))....(.....).........)))))).....))))..... (-24.09 = -24.65 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:21 2006