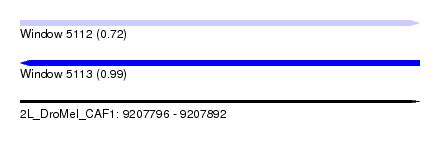
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,207,796 – 9,207,892 |
| Length | 96 |
| Max. P | 0.993545 |

| Location | 9,207,796 – 9,207,892 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -28.12 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
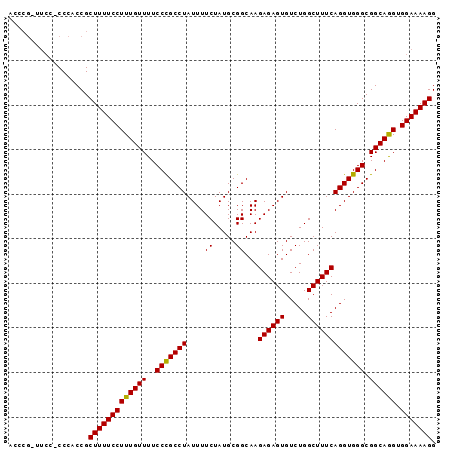
>2L_DroMel_CAF1 9207796 96 + 22407834 ACCCGUUUCCCCCCACCGCUUUUCCUUUGUUUUCCCGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAAGUGGAAAAGG ..((((((.((((((((.................((((.((.....)).))))...((((((.....)))))).)))))).)).))))))...... ( -29.70) >DroSec_CAF1 42959 95 + 1 ACCCGCUUCC-CCCGCCGCUUUUCCUUUGUUUUCCCGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAGGUGGAAAAGG ..((..((((-((.(((((((..(((........((((.((.....)).))))...((((((.....))))))))).))))))).)).))))..)) ( -35.70) >DroSim_CAF1 44754 96 + 1 ACCCGCUUCCUCCCACCGCUUUUCCUUUGUUUUCCCGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAGGUGGAAAAGG ..((((((((.((((((.................((((.((.....)).))))...((((((.....)))))).)))))).)).))))))...... ( -34.90) >DroEre_CAF1 48701 94 + 1 CCUCG--UCGUCCCUCCGCUUUUCCUUUGUUUUCCCGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAGGUGGAAAAGG (((..--((...(((((((((..(((........((((.((.....)).))))...((((((.....))))))))).)))))).)))..))..))) ( -29.00) >DroYak_CAF1 47758 87 + 1 AC---------CCCUCCGCUUUUCCUUUGUUUUCCUGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAGGUGGAAAAGG .(---------((((((((((..(((.........(((((((.....))).)))).((((((.....))))))))).)))))).))).))...... ( -29.10) >consensus ACCCG_UUCC_CCCACCGCUUUUCCUUUGUUUUCCCGCCUAUUUUCUAUGCGGCAAGAGAGUGUCUGGCUUUCAGGUGGGCGGCAGGUGGAAAAGG ..................(((((((((((((..(((((((....((.....))...((((((.....))))))))))))).)))))).))))))). (-28.12 = -27.80 + -0.32)


| Location | 9,207,796 – 9,207,892 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -26.70 |
| Energy contribution | -28.82 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
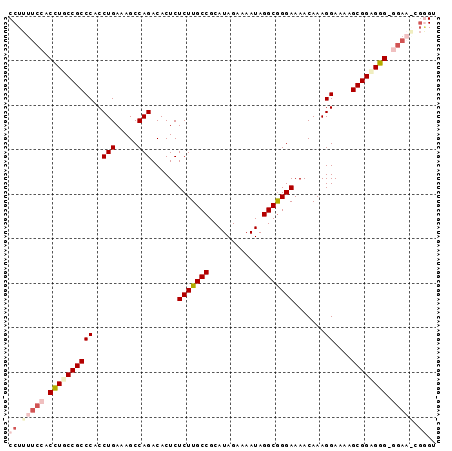
>2L_DroMel_CAF1 9207796 96 - 22407834 CCUUUUCCACUUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCGGGAAAACAAAGGAAAAGCGGUGGGGGGAAACGGGU ((.(((((.((..((((((..(((.....)))......(((((((............))))))).......))....))))..)).))))).)).. ( -33.20) >DroSec_CAF1 42959 95 - 1 CCUUUUCCACCUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCGGGAAAACAAAGGAAAAGCGGCGGG-GGAAGCGGGU ((.(((((.((((((((((..(((.....)))......(((((((............))))))).......))....))))))))-))))).)).. ( -37.00) >DroSim_CAF1 44754 96 - 1 CCUUUUCCACCUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCGGGAAAACAAAGGAAAAGCGGUGGGAGGAAGCGGGU ((.(((((.((..((((((..(((.....)))......(((((((............))))))).......))....))))..)).))))).)).. ( -34.60) >DroEre_CAF1 48701 94 - 1 CCUUUUCCACCUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCGGGAAAACAAAGGAAAAGCGGAGGGACGA--CGAGG ((((.((..(((.((((((..(((.....)))......(((((((............))))))).......))....)))))))...))--.)))) ( -26.80) >DroYak_CAF1 47758 87 - 1 CCUUUUCCACCUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCAGGAAAACAAAGGAAAAGCGGAGGG---------GU ......((.(((.((((((..(((.....)))......(((((((............))))))).......))....))))))))---------). ( -24.90) >consensus CCUUUUCCACCUGCCGCCCACCUGAAAGCCAGACACUCUCUUGCCGCAUAGAAAAUAGGCGGGAAAACAAAGGAAAAGCGGAGGG_GGAA_CGGGU ((.(((((.((((((((((..(((.....)))......(((((((............))))))).......))....)))))))).))))).)).. (-26.70 = -28.82 + 2.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:13 2006