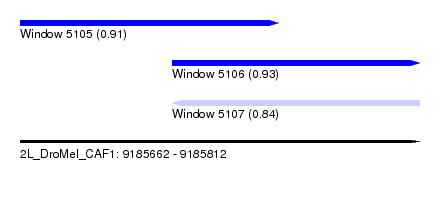
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,185,662 – 9,185,812 |
| Length | 150 |
| Max. P | 0.931803 |

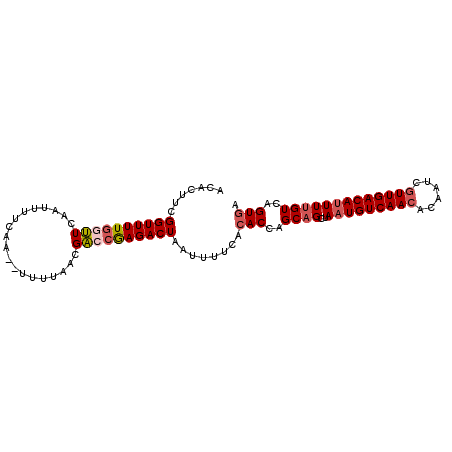
| Location | 9,185,662 – 9,185,759 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.15 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.51 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9185662 97 + 22407834 GCACUUCGGUUUUGGUUCAAUUUUCAA--UUUUAACGACCGAGACUAAUUUUCACACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGG .......((((((((((..........--.......))))))))))........(((..((((..(((((((((......)))))))))))))..))). ( -25.23) >DroSec_CAF1 21610 97 + 1 ACACUUCGGUUUUGCUUCAAUUUUCAA--UUUUAACGACCGAGACUAAUUUUUAAACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGA .((((((((((................--.......)))))).................((((..(((((((((......))))))))))))).)))). ( -19.40) >DroSim_CAF1 22487 97 + 1 ACACUUCGGUUUUGCUUCAAUUUUCAA--UUUUAACGACCGAGACUAAUUUUCACACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGA ....(((((((................--.......)))))))...........(((..((((..(((((((((......)))))))))))))..))). ( -21.10) >DroEre_CAF1 25324 97 + 1 ACACUUCGGUUUUGGUUCAGUUUUCAA--UUUUAACGACCGAGACUAAUUUUCACACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGA .......((((((((((..(((.....--....)))))))))))))........(((..((((..(((((((((......)))))))))))))..))). ( -26.20) >DroYak_CAF1 23503 97 + 1 ACAGUUCGGUUUUGGUUCAGUUUACAA--UUUUAACGACCGAGACUAAUUUUCACACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGA .......((((((((((..(((.....--....)))))))))))))........(((..((((..(((((((((......)))))))))))))..))). ( -26.20) >DroAna_CAF1 23454 97 + 1 A-ACUCUGGUUUUCGCUUA-UUUUCAACACUUUAAUGGCCAAGACUAAUUUUCACACCAGCGGCGAAUGUCAACAGGGCCAUUGACAGUUGGUCGGUGA .-..........(((((..-...(((((...(((((((((..(((.....(((.(.......).))).))).....)))))))))..)))))..))))) ( -21.70) >consensus ACACUUCGGUUUUGGUUCAAUUUUCAA__UUUUAACGACCGAGACUAAUUUUCACACCAGCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGA .......((((((((((...................))))))))))........(((..((((..(((((((((......)))))))))))))..))). (-19.76 = -20.51 + 0.75)

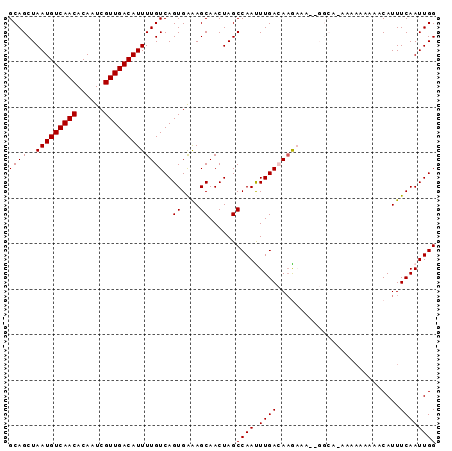
| Location | 9,185,719 – 9,185,812 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.86 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9185719 93 + 22407834 GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGGAAGCAACUAGCCAAUUUGACAAGAAAAAGGCA-AAAAAAAAACAUUUCAAUUGG ...(((..(((((((......)))))))((((((((.(((..........)))...))))))))....))).-..................... ( -21.10) >DroSec_CAF1 21667 83 + 1 GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGAGAGCAACUAGCCAAUUUGAAAAA----------AAGAAAA-ACAUUUCAAUUGG ((((..(((((((((......)))))))))))))..((....))......((((((.((((..----------.......-...)))))))))) ( -17.70) >DroSim_CAF1 22544 84 + 1 GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGAGAGCAACUAGCCAAUUUGAAAAA----------AAGAAAAUACAUUUCAAUUGG ((((..(((((((((......)))))))))))))..((....))......((((((.((((..----------...........)))))))))) ( -17.62) >DroEre_CAF1 25381 87 + 1 GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGAAAGCAACUAGCCAACUUGACAUGAAAA-GGCAAA------AACAUUUCAAUUGG ...((((((((((((......))))))))).(((((((....((.....))..)).)))))......-)))...------.............. ( -19.40) >DroYak_CAF1 23560 93 + 1 GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGAAAGCAACUAGCCAACUUGACAAGAAAU-GGCACAAAAAAACACAUUUCAAUUGG ...((((.(((((((......)))))))((((((((((....((.....))..)).))))))))..)-)))....................... ( -22.20) >consensus GCAGCUAAUGUCAACACAAUCGUUGACAUUUUGUCAGUGAAAGCAACUAGCCAAUUUGACAAGAAA__GGCA_AAAAAAAAACAUUUCAAUUGG ....(((((((((((......)))))).(((((((((.....((.....))....))))))))).........................))))) (-14.10 = -14.86 + 0.76)

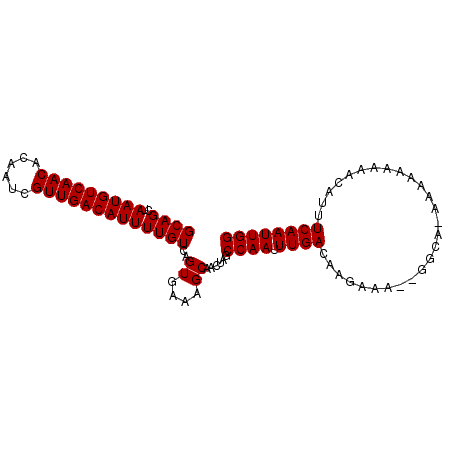
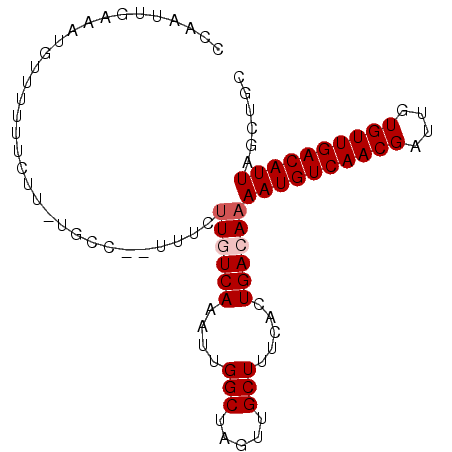
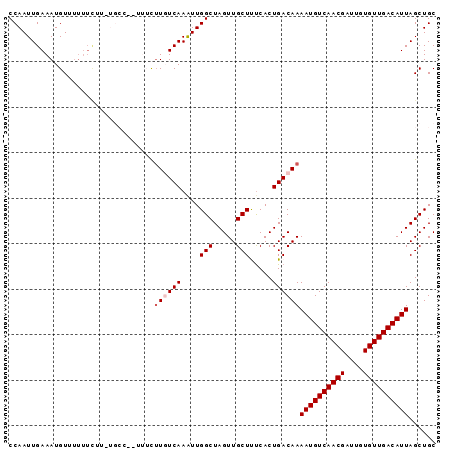
| Location | 9,185,719 – 9,185,812 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9185719 93 - 22407834 CCAAUUGAAAUGUUUUUUUUU-UGCCUUUUUCUUGUCAAAUUGGCUAGUUGCUUCCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC .(((..((((....))))..)-))........((((((....(((.....))).....))))))((((((((((....))))))))))...... ( -20.40) >DroSec_CAF1 21667 83 - 1 CCAAUUGAAAUGU-UUUUCUU----------UUUUUCAAAUUGGCUAGUUGCUCUCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC .(((((((((...-.......----------..)))).)))))(((..(((.((.....)))))((((((((((....)))))))))))))... ( -17.20) >DroSim_CAF1 22544 84 - 1 CCAAUUGAAAUGUAUUUUCUU----------UUUUUCAAAUUGGCUAGUUGCUCUCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC .(((((((((...........----------..)))).)))))(((..(((.((.....)))))((((((((((....)))))))))))))... ( -17.12) >DroEre_CAF1 25381 87 - 1 CCAAUUGAAAUGUU------UUUGCC-UUUUCAUGUCAAGUUGGCUAGUUGCUUUCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC .....(((((.((.------...)).-.)))))(((((.((.(((.....)))...))))))).((((((((((....))))))))))...... ( -21.10) >DroYak_CAF1 23560 93 - 1 CCAAUUGAAAUGUGUUUUUUUGUGCC-AUUUCUUGUCAAGUUGGCUAGUUGCUUUCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC ......((((((.((........)))-)))))((((((.((.(((.....)))...))))))))((((((((((....))))))))))...... ( -25.40) >consensus CCAAUUGAAAUGUUUUUUCUU_UGCC__UUUCUUGUCAAAUUGGCUAGUUGCUUUCACUGACAAAAUGUCAACGAUUGUGUUGACAUUAGCUGC ................................((((((....(((.....))).....))))))((((((((((....))))))))))...... (-15.98 = -16.58 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:07 2006