
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,184,493 – 9,184,620 |
| Length | 127 |
| Max. P | 0.993071 |

| Location | 9,184,493 – 9,184,585 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -12.35 |
| Energy contribution | -15.53 |
| Covariance contribution | 3.18 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9184493 92 + 22407834 GCCAUUGCAAAUGGAUUCGGAUUCGUUUGUGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUA---------UAUAUACAUAUAAAGCAUCCGCAUUAUGCAG .....((((....(((.(((((.(.((((((((.(((-----(((((..((........))..)))---------))))).))))))))).))))).))).)))). ( -24.80) >DroSec_CAF1 20447 101 + 1 GCCAUUGCGAAUGGAUUCGGAUUCGUUUGUGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUUCGCAUUAUGCAG ((...((((((((..............((((((((((-----(.........)))))))))))((((((((...))))))))........))))))))....)).. ( -29.20) >DroSim_CAF1 21304 101 + 1 GCCAUUGCAAAUGGAUUCGGAUUCGUUUGCGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAG ......((((((((((....))))))))))(((((((-----(.........))))))))...((((((((...)))))))).(((((.((....)).)))))... ( -25.90) >DroEre_CAF1 24140 106 + 1 GCCAUUGCAAAUGGAUUCGGAUUCGUUUGCGUGCAUAUAGUAUAUAUCUUGAAUAUGUACAUCGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAG ......((((((((((....)))))))))).((((((..((((((((((...((((((((...)))))))))).)))))))).......((....))..)))))). ( -29.50) >DroAna_CAF1 22434 85 + 1 UCUAUUGCCAGUGGACUGC-----GAGUGUGUGAAUA-----CAUUGCCUGGCCACGUACAUAGUU-----------UAUUCAUAUAAAGCGUCCAAAUUAUGCAG .....(((...(((((.((-----...((((((((((-----.((((..((........)))))).-----------))))))))))..)))))))......))). ( -22.70) >consensus GCCAUUGCAAAUGGAUUCGGAUUCGUUUGUGUGCAUA_____UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAG ......((((((((((....))))))))))(((((((................)))))))...(((((((.....))))))).......((((.......)))).. (-12.35 = -15.53 + 3.18)

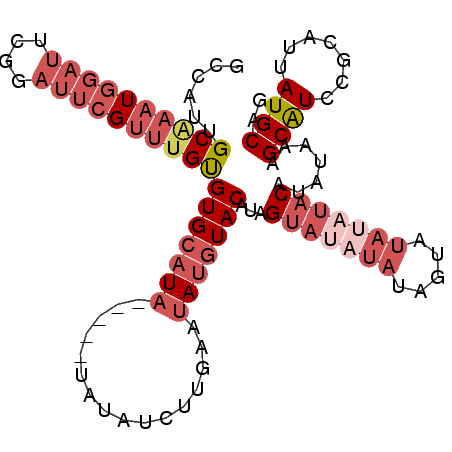

| Location | 9,184,522 – 9,184,620 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9184522 98 + 22407834 UGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUA---------UAUAUACAUAUAAAGCAUCCGCAUUAUGCAGAAAGAAACUGGCUAGAAUCGAAAACAGUUUGGGUA ((((.(((-----(((((..((........))..)))---------))))).))))......((((((.....))......((((((..............)))))))))). ( -18.24) >DroSec_CAF1 20476 107 + 1 UGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUUCGCAUUAUGCAGAAAGAAACUGGCUAGAAUCGAAAACAGUUUGGGCA ((((((((-----(.........)))))))))..((((((((...)))))))).......((.(((((.....)).)))..((((((..............))))))..)). ( -21.54) >DroSim_CAF1 21333 107 + 1 CGUGCAUA-----UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAGAAAGAAACUGGCUAGAAUCGAAAACAGUUUGGGCA .(((((((-----(.........))))))))...((((((((...)))))))).......((.((.((.....)).))...((((((..............))))))..)). ( -21.44) >DroEre_CAF1 24169 112 + 1 CGUGCAUAUAGUAUAUAUCUUGAAUAUGUACAUCGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAGAAAGAAACUGACUACAAUCGAAAACAGUUUGGGAA ..(((.((((((((((((((...((((((((...)))))))))).)))))))).))))..)))(((((.....))......((((((..............)))))).))). ( -22.24) >consensus CGUGCAUA_____UAUAUCUUGAAUAUGUACAUAGUAUAUAUAGUAUAUAUACAUAUAAAGCAUCCGCAUUAUGCAGAAAGAAACUGGCUAGAAUCGAAAACAGUUUGGGCA .(((((((................)))))))...(((((((.....)))))))..........(((((.....))......((((((..............))))))))).. (-15.17 = -15.98 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:05 2006