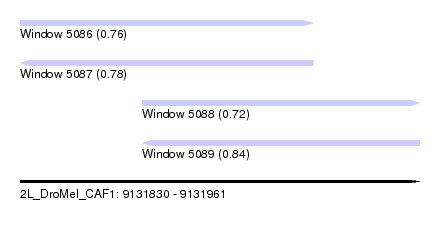
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,131,830 – 9,131,961 |
| Length | 131 |
| Max. P | 0.844038 |

| Location | 9,131,830 – 9,131,926 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 74.64 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -10.78 |
| Energy contribution | -10.87 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.762014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
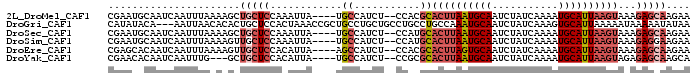
>2L_DroMel_CAF1 9131830 96 + 22407834 AAAAAUUCCAUUGCCUUGGAGUGCAAAACAGCUGCAAUAUUUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGUGG--AGAUGGC ....((((((......))))))(((.......)))..((((((((((.....((((((((((.((.....)).))))))))))))).))--))))).. ( -25.10) >DroGri_CAF1 29530 90 + 1 -A--AAUGCA-----UCCAGCUGCGGAAGGGGGUCAAUUCUUAUAUUUUUAUUUUUAAUGCACUUUGAUAGAUUGCAUUUGGCAGGCAGGCAGCAGGC -.--..(((.-----.((.(((((.(((.(.((((...((........(((....)))........))..)))).).))).))).)).))..)))... ( -16.59) >DroSec_CAF1 19328 86 + 1 GAAAAUUCCAUU----------GCAAAACAGCUGAAAUAUUUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCAUGG--AGAUGGC .....((((((.----------((.(((.(((.(((....)))..))).)))((((((((((.((.....)).))))))))))))))))--))..... ( -23.40) >DroSim_CAF1 19628 96 + 1 AAAAAUUCCAUUGCCUUCCAGUGCAAAACAGCUGCAAUAUUUCUUCCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCAUGG--AGAUGGC ............((((((((.((((.......))))...............(((((((((((.((.....)).)))))))))))..)))--))..))) ( -22.90) >DroEre_CAF1 19663 96 + 1 AAAAAUUACACUGCCUUCCAGUGCACAACAGCUGCAAUAUUUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCACUAAGUGCGUGG--AGAUGGC ............((((((((.(((((...((.((((((...((.(((............)))....))...))))))))..))))))))--))..))) ( -24.60) >DroYak_CAF1 20016 85 + 1 AAAAAUUGCAGUGCUUUCCAGUG-----------CAAUAUUGCUUGCUCUCUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGCGG--AGAUGGC ....((((((.((.....)).))-----------))))...(((..((((((((((((((((.((.....)).))))))))))).).))--))..))) ( -26.30) >consensus AAAAAUUCCAUUGCCUUCCAGUGCAAAACAGCUGCAAUAUUUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGUGG__AGAUGGC ...................................................(((((((((((.((.....)).))))))))))).............. (-10.78 = -10.87 + 0.08)

| Location | 9,131,830 – 9,131,926 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 74.64 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -8.52 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9131830 96 - 22407834 GCCAUCU--CCACGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAAAUAUUGCAGCUGUUUUGCACUCCAAGGCAAUGGAAUUUUU ......(--(((.((((((((((((...........)))))))))).....((((......)))).))(((((((.....))))))).))))...... ( -24.60) >DroGri_CAF1 29530 90 - 1 GCCUGCUGCCUGCCUGCCAAAUGCAAUCUAUCAAAGUGCAUUAAAAAUAAAAAUAUAAGAAUUGACCCCCUUCCGCAGCUGGA-----UGCAUU--U- .((.(((((..........((((((...........))))))................(((.........))).))))).)).-----......--.- ( -13.70) >DroSec_CAF1 19328 86 - 1 GCCAUCU--CCAUGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAAAUAUUUCAGCUGUUUUGC----------AAUGGAAUUUUC ......(--((((((((((((((((...........))))))))))(((.(((..(((....))).))).))).))----------.)))))...... ( -21.50) >DroSim_CAF1 19628 96 - 1 GCCAUCU--CCAUGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGGAAGAAAUAUUGCAGCUGUUUUGCACUGGAAGGCAAUGGAAUUUUU (((...(--((((((((((((((((...........)))))))))).........((((((.......))))))))).)))).)))............ ( -23.40) >DroEre_CAF1 19663 96 - 1 GCCAUCU--CCACGCACUUAGUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAAAUAUUGCAGCUGUUGUGCACUGGAAGGCAGUGUAAUUUUU .......--....((((((((((((...........)))))))))).....((((......)))).)).....(((((((.....)))))))...... ( -24.80) >DroYak_CAF1 20016 85 - 1 GCCAUCU--CCGCGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAGAGAGCAAGCAAUAUUG-----------CACUGGAAAGCACUGCAAUUUUU ((..(((--(.....((((((((((...........)))))))))).))))....))...((((-----------((.((.....)).)))))).... ( -20.10) >consensus GCCAUCU__CCACGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAAAUAUUGCAGCUGUUUUGCACUGGAAGGCAAUGGAAUUUUU ...............((((((((((...........)))))))))).................................................... ( -8.52 = -9.05 + 0.53)


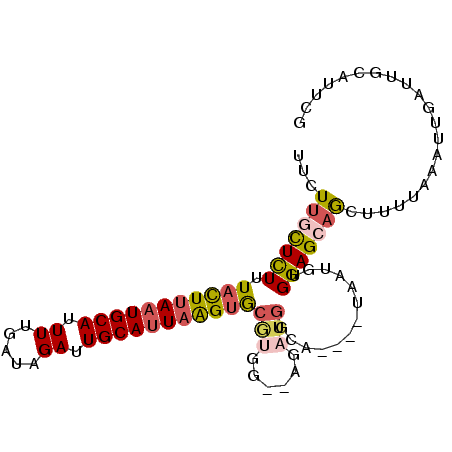
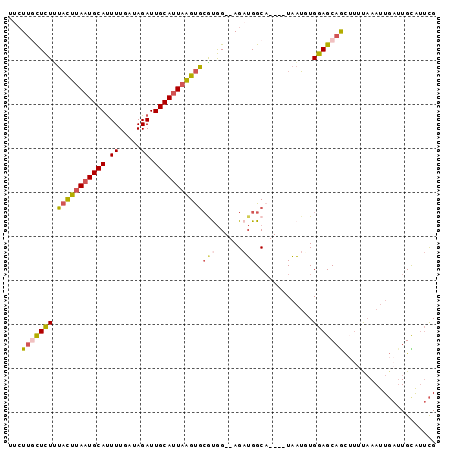
| Location | 9,131,870 – 9,131,961 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9131870 91 + 22407834 UUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGUGG--AGAUGGCA----UAAUUUGGAGCAGCUUUUAAAUUGAUUGCAUUCG ......((((.(((((((((((.((.....)).)))))))))))...))--))...(((----((((((((((....))))))))))..)))..... ( -22.90) >DroGri_CAF1 29562 94 + 1 UUAUAUUUUUAUUUUUAAUGCACUUUGAUAGAUUGCAUUUGGCAGGCAGGCAGCAGGCAGCGGUUUAGUGGAGCAGUGUGUUAAUU---UGUAUAUG .................(((((..((((((.(((((.....((..((.....))..)).((......))...))))).))))))..---)))))... ( -21.50) >DroSec_CAF1 19358 91 + 1 UUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCAUGG--AGAUGGCA----UAAUUUGGAGCAGCUUUUAAAUUGAUUGCAUUCG ......((((.(((((((((((.((.....)).)))))))))))...))--))...(((----((((((((((....))))))))))..)))..... ( -22.90) >DroSim_CAF1 19668 91 + 1 UUCUUCCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCAUGG--AGAUGGCA----UAAUUUGGAGCAACUUUUAAAUUGAUUGCAUUCG ......((((.(((((((((((.((.....)).)))))))))))...))--))...(((----((((((((((....))))))))))..)))..... ( -23.00) >DroEre_CAF1 19703 91 + 1 UUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCACUAAGUGCGUGG--AGAUGGCU----UAAUGUGGAGCAACUUUUAAAUUGAUUGUGCUCG ......((((.((((((.((((.((.....)).)))).))))))...))--))......----.......(((((...((......))...))))). ( -19.80) >DroYak_CAF1 20045 88 + 1 UGCUUGCUCUCUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGCGG--AGAUGGCA----UAAUGUGGAGCAGC---CAAAUUGAUUGUGUUCG ((((..((((((((((((((((.((.....)).))))))))))).).))--))..))))----.......(((((..---(((.....)))))))). ( -24.50) >consensus UUCUUGCUCUUUACUUAAUGCAUUUUGAUAGAUUGCAUUAAGUGCGUGG__AGAUGGCA____UAAUGUGGAGCAGCUUUUAAAUUGAUUGCAUUCG ...(((((((.(((((((((((.((.....)).)))))))))))(((......))).............)))))))..................... (-14.36 = -14.95 + 0.59)

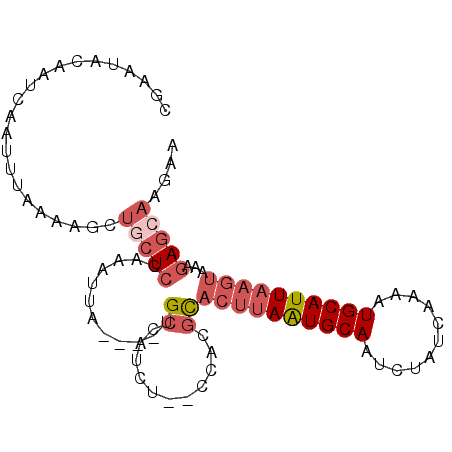
| Location | 9,131,870 – 9,131,961 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -16.02 |
| Consensus MFE | -9.98 |
| Energy contribution | -11.37 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9131870 91 - 22407834 CGAAUGCAAUCAAUUUAAAAGCUGCUCCAAAUUA----UGCCAUCU--CCACGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAA ......................(((((.......----.((.....--....))((((((((((...........))))))))))...))))).... ( -16.70) >DroGri_CAF1 29562 94 - 1 CAUAUACA---AAUUAACACACUGCUCCACUAAACCGCUGCCUGCUGCCUGCCUGCCAAAUGCAAUCUAUCAAAGUGCAUUAAAAAUAAAAAUAUAA ........---..((((..((((.............((.((.....))..)).(((.....))).........))))..)))).............. ( -7.30) >DroSec_CAF1 19358 91 - 1 CGAAUGCAAUCAAUUUAAAAGCUGCUCCAAAUUA----UGCCAUCU--CCAUGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAA ......................(((((.......----.((.....--....))((((((((((...........))))))))))...))))).... ( -16.60) >DroSim_CAF1 19668 91 - 1 CGAAUGCAAUCAAUUUAAAAGUUGCUCCAAAUUA----UGCCAUCU--CCAUGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGGAAGAA .....(((((..........))))).........----.....(((--((.(..((((((((((...........))))))))))..)..)).))). ( -16.00) >DroEre_CAF1 19703 91 - 1 CGAGCACAAUCAAUUUAAAAGUUGCUCCACAUUA----AGCCAUCU--CCACGCACUUAGUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAA .((......))..........((((((.......----.((.....--....))((((((((((...........))))))))))...))))))... ( -18.00) >DroYak_CAF1 20045 88 - 1 CGAACACAAUCAAUUUG---GCUGCUCCACAUUA----UGCCAUCU--CCGCGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAGAGAGCAAGCA ......(((.....)))---((((((((......----.(((....--..).))((((((((((...........)))))))))).).)))).))). ( -21.50) >consensus CGAAUACAAUCAAUUUAAAAGCUGCUCCAAAUUA____UGCCAUCU__CCACGCACUUAAUGCAAUCUAUCAAAAUGCAUUAAGUAAAGAGCAAGAA ......................(((((............((...........))((((((((((...........))))))))))...))))).... ( -9.98 = -11.37 + 1.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:50 2006