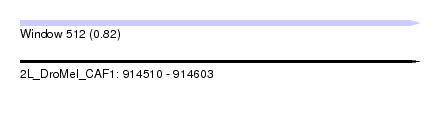
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 914,510 – 914,603 |
| Length | 93 |
| Max. P | 0.818618 |

| Location | 914,510 – 914,603 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
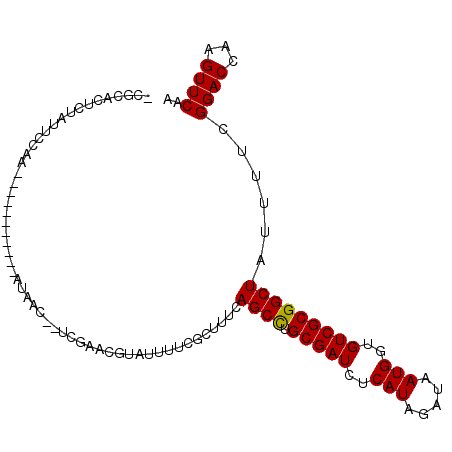
>2L_DroMel_CAF1 914510 93 + 22407834 -CGCACUCUAUUCCAA-----------AUAAC--UCGAACAUAUUUUCGCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA -...............-----------.....--..((((.....((((.....((((.(((((..(((.....)))..))))))))).....))))....)))).. ( -20.90) >DroSec_CAF1 43624 93 + 1 -CGCACUCUAUUCCAA-----------AUCAC--UCGAACGUAUUUUCGCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA -...............-----------.....--..((((.....((((.....((((.(((((..(((.....)))..))))))))).....))))....)))).. ( -22.00) >DroSim_CAF1 41500 93 + 1 -CGCACUCUAUUCCAA-----------AUCAC--UCGAACGUAUUUUCGCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA -...............-----------.....--..((((.....((((.....((((.(((((..(((.....)))..))))))))).....))))....)))).. ( -22.00) >DroEre_CAF1 50557 100 + 1 ----GCAAU-UUCCAAACUCAAUCCAAAUGAU--UCCAUCGAAUUUUCGCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA ----.....-.....((((...(((....(((--((....))))).........((((.(((((..(((.....)))..)))))))))......)))...))))... ( -21.10) >DroYak_CAF1 45220 100 + 1 ----UCUCUAUUCCAAAUAAAAUCGAACGUAU---UUUCCGAAUUUUCCCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA ----....................((((....---..((((((..(........((((.(((((..(((.....)))..))))))))))..))))))....)))).. ( -24.50) >DroPer_CAF1 42271 96 + 1 UCGCAUUCUAUUCGAG-----------UGAAUUUUCAAUUUCGAUUUCGCUUUCAGCUUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA ..........((((((-----------((((...((......)).)))))....((((.(((((..(((.....)))..)))))))))....))))).......... ( -21.50) >consensus _CGCACUCUAUUCCAA___________AUAAC__UCGAACGUAUUUUCGCUUUCAGCCUGCGAUCUCAUAGAUAAUGGUGUCGCGGCUAUUUUCGGACCAAGUUCAA ......................................................((((.(((((..(((.....)))..)))))))))......((((...)))).. (-16.57 = -16.43 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:58 2006