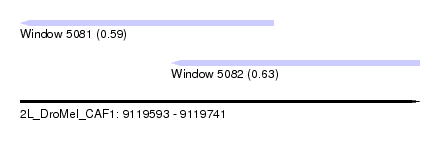
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,119,593 – 9,119,741 |
| Length | 148 |
| Max. P | 0.626871 |

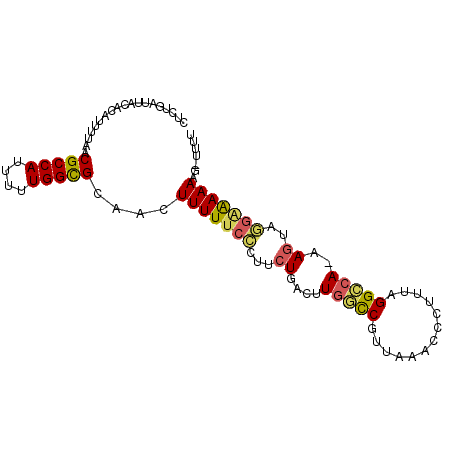
| Location | 9,119,593 – 9,119,687 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.14 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
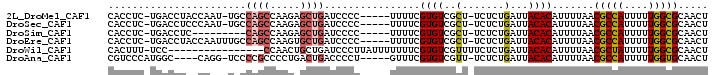
>2L_DroMel_CAF1 9119593 94 - 22407834 CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUUCCCUUCUGACUUGGCCGUUAAACCCUUUAGGCCA-AAGUCGGAAAAAAG-UUUU ...................(((((....))))).((((((((....((((((((((((.............)))).-))))))))))))))-)).. ( -28.92) >DroVir_CAF1 10048 86 - 1 CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUGC-----UGAGCUACUCGUAAAACCCUUUAGGCCA-GACCAG--AAAAAG-UUU- ...................(((((....))))).(((((((.(-----((..((...(.((((....)))).)..)-)..)))--.)))))-)).- ( -16.80) >DroSec_CAF1 7128 94 - 1 CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUUCCCUUCUGACUUGGUCGUUAAACCCUUUAGGCCA-AAGUAGGAAAAAAG-UUUU ...................(((((....))))).((((((((.(((.((...((((((.............)))))-))).))).))))))-)).. ( -23.12) >DroEre_CAF1 7222 94 - 1 CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUUCCCCUCUGACUUGGCCGUUAAACCCUUUAGGCCA-GAGUAGGAAAAAGU-UGUU ....................((((....))))((((((((((((.((((.....((((.............)))))-)))..)))))))))-))). ( -30.32) >DroYak_CAF1 7420 95 - 1 CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUUCCCUUCUGACGUGGCCGUUAAACCCUUUAGGCCAAAAGUAGGAAAAAAG-UUUU ...................(((((....))))).((((((((.(((.((....(((((.............)))))..)).))).))))))-)).. ( -26.92) >DroAna_CAF1 23418 96 - 1 CUCUGAUUACACAUUUUAACGCCAUUUUUGGUGCAACUUUUUCUCUUCUGACUUGCCCGUUAACCCCUUUAGGGCAACAGAAAGGGAAAAGUUUUU ...................(((((....))))).(((((((((..(((((..((((((.............)))))))))))..)))))))))... ( -29.72) >consensus CUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACUUUUUCCCUUCUGACUUGGCCGUUAAACCCUUUAGGCCA_AAGUAGGAAAAAAG_UUUU ...................(((((....)))))....(((((((...((....(((((.............)))))..))..)))))))....... (-17.19 = -17.14 + -0.05)


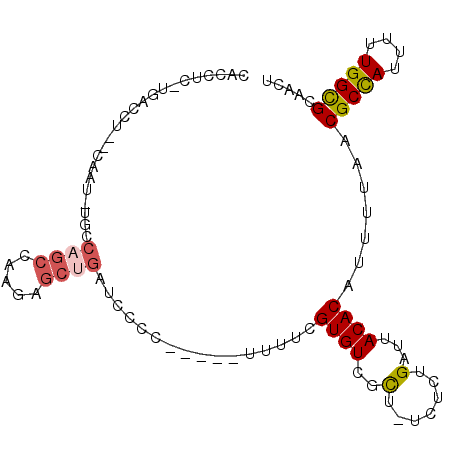
| Location | 9,119,649 – 9,119,741 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -16.08 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9119649 92 - 22407834 CACCUC-UGACCUACCAAU-UGCCAGCCAAGAGCUGAUCCCC-----UUUUCGUGUCGCU-UCUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACU ......-...........(-(((((((.....))))......-----.....((((..(.-.....)...))))........((((....)))))))).. ( -16.70) >DroSec_CAF1 7184 92 - 1 CACCUC-UGACCUCCCAAU-UGCCAGCCAAGAGCUGAUCCCC-----UUUUCGUGUCGCU-UCUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACU ......-...........(-(((((((.....))))......-----.....((((..(.-.....)...))))........((((....)))))))).. ( -16.70) >DroSim_CAF1 7222 84 - 1 CACCUC-UGACCUC---------CAGCCAAGAGCUGAUCCCC-----UUUUCGUGUCGCU-UCUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACU ......-.......---------((((.....))))......-----.....((((..(.-.....)...)))).......(((((....)))))..... ( -15.20) >DroEre_CAF1 7278 93 - 1 CACCUC-UGACCUACCAAUUUGCCAGCCAAGUGCUGAUCCCC-----UUUUCGUGUCGCU-UCUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACU ......-............(((((((..(((((((((.....-----...))).).))))-)..)))...............((((....)))))))).. ( -17.20) >DroWil_CAF1 34721 83 - 1 CACUUU-UCC----------------CCAACUGCUGAUCCCUUAUUUUUUUCGUGUCGUUUUCUCUGAUUACACAUUUUAACGCUAUUUUUGGCGCAACU ......-...----------------.....(((..................((((.(((......))).))))........((((....)))))))... ( -11.70) >DroAna_CAF1 23476 89 - 1 CGUCCCAUGGC----CAGG-UCCCCGCCCCUGACUGACCCCU-----GUUUCGUGUCGUU-UCUCUGAUUACACAUUUUAACGCCAUUUUUGGUGCAACU .(..(((((((----((((-........))))..........-----.....((((.(((-.....))).))))........))))....)))..).... ( -19.00) >consensus CACCUC_UGACCU__CAAU_UGCCAGCCAAGAGCUGAUCCCC_____UUUUCGUGUCGCU_UCUCUGAUUACACAUUUUAACGCCAUUUUUGGCGCAACU .......................((((.....))))................((((..(.......)...)))).......(((((....)))))..... ( -9.90 = -10.23 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:44 2006