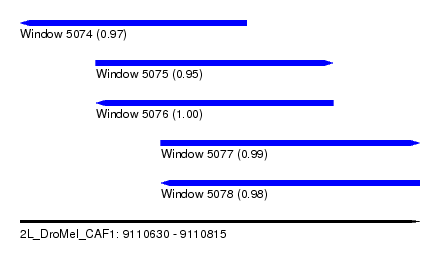
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,110,630 – 9,110,815 |
| Length | 185 |
| Max. P | 0.998994 |

| Location | 9,110,630 – 9,110,735 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 98.10 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110630 105 - 22407834 UUACGUUGGCCAAUUAUAAUUUUAAUUGUUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAAAGACAAUGUCUGAAAAUUACCACCACUUGCUGGGA .((((((.(.((((((......))))))..).))))))(((.........))).........((((((.......))))))........(((.(....).))).. ( -18.80) >DroSec_CAF1 12741 105 - 1 UUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAUGCAUAAAGACAAUGUCUGAAAAUUACCACCACUUGCUGGGA .(((((..((((((((......))))))))..).))))((.(((.....((....))....)))))....((((...))))........(((.(....).))).. ( -22.60) >DroSim_CAF1 12869 105 - 1 UUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAAAGACAAUGUCUGAAAAUUACCACCACUUGCUGGGA .(((((..((((((((......))))))))..).))))(((.........))).........((((((.......))))))........(((.(....).))).. ( -25.40) >DroEre_CAF1 12839 105 - 1 UUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAAAGACAAUGUCGGAAAAUUACCAGCACUUGCUGGGA .(((((..((((((((......))))))))..).)))).((........((....))......(((((.......))))))).......(((((....))))).. ( -31.80) >DroYak_CAF1 17808 105 - 1 UUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAAAGACAAUGUCUGAAAAUAACCACCACUUGCUGGGA .(((((..((((((((......))))))))..).))))(((.........))).........((((((.......))))))........(((.(....).))).. ( -25.40) >consensus UUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAAAGACAAUGUCUGAAAAUUACCACCACUUGCUGGGA .(((((..((((((((......))))))))..).))))(((.........))).........((((((.......))))))........(((.(....).))).. (-22.30 = -22.90 + 0.60)

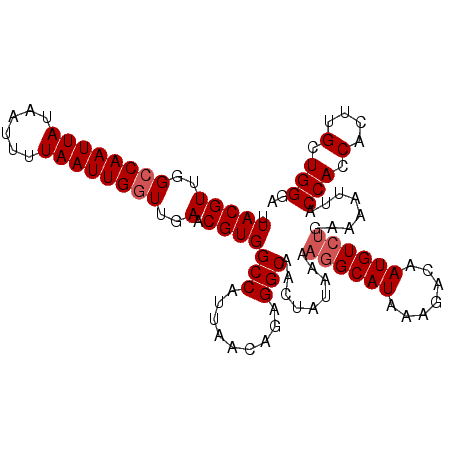
| Location | 9,110,665 – 9,110,775 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -18.50 |
| Energy contribution | -19.74 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110665 110 + 22407834 UUAUGCCUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAAACAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCAGCCAGAGAAAAUC ...................(((((....((((.(((((....((((((......))))))...)))))....((((.............))))))))..)))))...... ( -22.12) >DroSec_CAF1 12776 110 + 1 UUAUGCAUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCUUCCAGAGAAAAUC ....(((.........)))(((((.....(((.(((((...(((((((......)))))))..)))))....((((.............)))))))...)))))...... ( -23.62) >DroSim_CAF1 12904 110 + 1 UUAUGCCUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUACCGCCUUCCAGAGAAAAUC ...................(((((.....(((.(((((...(((((((......)))))))..)))))....((.....))............)))...)))))...... ( -21.20) >DroEre_CAF1 12874 110 + 1 UUAUGCCUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACGUAGCGCCAUCCAAAGAAAAUC ....................(((....(((((.(((((...(((((((......)))))))..)))))....((((.............)))))))))....)))..... ( -20.62) >DroYak_CAF1 17843 110 + 1 UUAUGCCUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACGUAGCGCCAUCCAAGGAAAAUC .....((((..((((......))))..(((((.(((((...(((((((......)))))))..)))))....((((.............)))))))))..))))...... ( -21.52) >consensus UUAUGCCUUUUAUAGUUGCCUCUGUUAAUGGCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCAUCCAGAGAAAAUC ...................(((((...(((((.(((((...(((((((......)))))))..)))))....((((.............))))))))).)))))...... (-18.50 = -19.74 + 1.24)

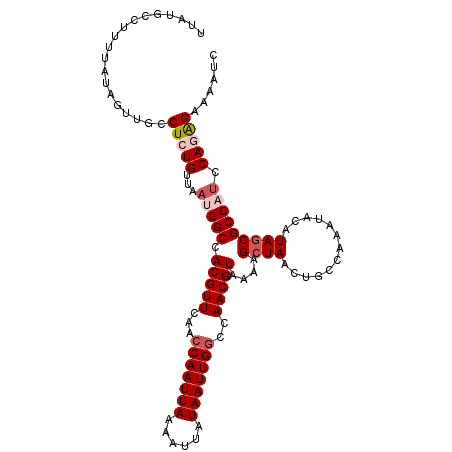

| Location | 9,110,665 – 9,110,775 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110665 110 - 22407834 GAUUUUCUCUGGCUGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGUUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAA .....((((((((((((((((......)))).))))))........((((((...(((((....))))).......))))))....)))))).................. ( -28.60) >DroSec_CAF1 12776 110 - 1 GAUUUUCUCUGGAAGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAUGCAUAA ......(((((...(((((((......))))..........(((((..((((((((......))))))))..).))))))).....)))))(((.........))).... ( -32.20) >DroSim_CAF1 12904 110 - 1 GAUUUUCUCUGGAAGGCGGUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAA .....((((((((((((....(((.....)))....))))))((((..((((((((......))))))))..).))).........)))))).................. ( -29.40) >DroEre_CAF1 12874 110 - 1 GAUUUUCUUUGGAUGGCGCUACGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAA .....((((((((((((((((......))))..........(((((..((((((((......))))))))..).))))))))))..)))))).................. ( -31.30) >DroYak_CAF1 17843 110 - 1 GAUUUUCCUUGGAUGGCGCUACGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAA ......((((.((((((((((......))))..........(((((..((((((((......))))))))..).)))))))))).....((....))....))))..... ( -32.10) >consensus GAUUUUCUCUGGAUGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGCCAUUAACAGAGGCAACUAUAAAAGGCAUAA .....((((((((((((((((......))))..........(((((..((((((((......))))))))..).))))))))))..)))))).................. (-27.74 = -28.70 + 0.96)

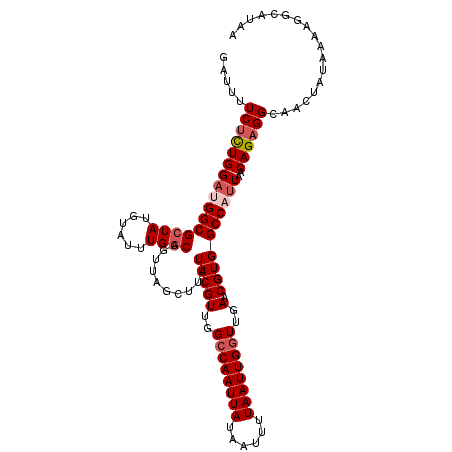
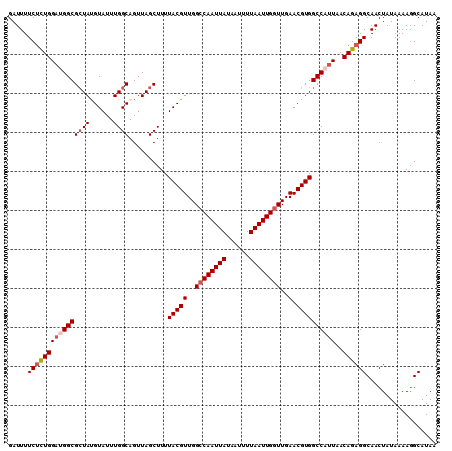
| Location | 9,110,695 – 9,110,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110695 120 + 22407834 GCCACGUUCAAACAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCAGCCAGAGAAAAUCUCUCUUUAAGCCCUGUUAAGUUUUCGCAGUGCACCCGACU ...(((((....((((((......))))))...)))))....((..(((((.(((.((.(((((...((.(((((.....)))))....))..))))).))))).)))))))........ ( -23.90) >DroSec_CAF1 12806 120 + 1 GCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCUUCCAGAGAAAAUCUCUCUUUAAGCCCUGUUAAGUUUUCGCAGUGCACCCGACU ...(((((...(((((((......)))))))..)))))....((..(((((.(((.((.(((((.(((..(((((.....)))))..)))...))))).))))).)))))))........ ( -27.20) >DroSim_CAF1 12934 120 + 1 GCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUACCGCCUUCCAGAGAAAAUCUCUCUUUAAGCCCUGUUAAGUUUUCGCAGUGCACCCGACU ...(((((...(((((((......)))))))..)))))....((..(((((.(((.....((.(.(((..(((((.....)))))..))).)..)).....))).)))))))........ ( -23.70) >DroEre_CAF1 12904 119 + 1 GCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACGUAGCGCCAUCCAAAGAAAAUCUC-CUUUAAGCCCUGCUAACUUUUCGCAGUGCAUCCGACU ...(((((...(((((((......)))))))..)))))....((..(((((.(((....(((((......((((........-))))......)))))...))).)))))))........ ( -23.80) >DroYak_CAF1 17873 120 + 1 GCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACGUAGCGCCAUCCAAGGAAAAUCUCUCUUUAAGCCCUGUUAACUUUUCGCAGUGCAGCCGACU ...(((((...(((((((......)))))))..)))))....(((.(((((.(((....(((((......(((((.......)))))......)))))...))).)))))..)))..... ( -24.60) >consensus GCCACGUUCAACCAAUUAAAAUUAUAAUUGGCCAACGUAAAAGCUAACUGCCAAAUACAUAGCGCCAUCCAGAGAAAAUCUCUCUUUAAGCCCUGUUAAGUUUUCGCAGUGCACCCGACU ...(((((...(((((((......)))))))..)))))....((..(((((.(((...((((.((.....(((((.....)))))....)).)))).....))).)))))))........ (-21.64 = -22.12 + 0.48)



| Location | 9,110,695 – 9,110,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.76 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110695 120 - 22407834 AGUCGGGUGCACUGCGAAAACUUAACAGGGCUUAAAGAGAGAUUUUCUCUGGCUGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGUUUGAACGUGGC .((((((((((..(((....((....))((((...((((((...))))))))))..)))..)))))))))).........(((((((.(.((((((......))))))..).))))))). ( -31.50) >DroSec_CAF1 12806 120 - 1 AGUCGGGUGCACUGCGAAAACUUAACAGGGCUUAAAGAGAGAUUUUCUCUGGAAGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGC .((((((((((..((.....((....))(.(((..((((((...))))))..))).)))..)))))))))).........((((((..((((((((......))))))))..).))))). ( -38.50) >DroSim_CAF1 12934 120 - 1 AGUCGGGUGCACUGCGAAAACUUAACAGGGCUUAAAGAGAGAUUUUCUCUGGAAGGCGGUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGC .((((((((((((((.....((.....)).(((..((((((...))))))..))))))))..))))))))).........((((((..((((((((......))))))))..).))))). ( -37.60) >DroEre_CAF1 12904 119 - 1 AGUCGGAUGCACUGCGAAAAGUUAGCAGGGCUUAAAG-GAGAUUUUCUUUGGAUGGCGCUACGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGC .(((((((((.((((.........)))).(((....(-(((.....))))....))).....))))))))).........((((((..((((((((......))))))))..).))))). ( -37.40) >DroYak_CAF1 17873 120 - 1 AGUCGGCUGCACUGCGAAAAGUUAACAGGGCUUAAAGAGAGAUUUUCCUUGGAUGGCGCUACGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGC (((..(((((..((((....((((.(((((...(((......))).)))))..))))....))))....)))))..))).((((((..((((((((......))))))))..).))))). ( -37.40) >consensus AGUCGGGUGCACUGCGAAAACUUAACAGGGCUUAAAGAGAGAUUUUCUCUGGAUGGCGCUAUGUAUUUGGCAGUUAGCUUUUACGUUGGCCAAUUAUAAUUUUAAUUGGUUGAACGUGGC .(((((((((.(((...........))).(((...((((((...))))))....))).....))))))))).........((((((..((((((((......))))))))..).))))). (-30.32 = -30.76 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:40 2006