
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,110,033 – 9,110,220 |
| Length | 187 |
| Max. P | 0.999884 |

| Location | 9,110,033 – 9,110,142 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110033 109 + 22407834 GAACAGAAUUUAAUAAAACCUAUUUCCAUUUAUUAAAUUCUUCUCUUCAUCGUCGCCUCUAAUU-UUGCUUUGCCACAAACUUUGGCCGGUUUUCCCGACAGCCGUGGCA ((..(((((((((((((...........)))))))))))))..)).........(((.......-.......((((.......))))(((((........))))).))). ( -22.80) >DroSec_CAF1 12149 109 + 1 GAACAGAAUUUAAUAAAACCUUCUUCCAUUUAUUAAAUUCUUCUCUUCAUCGUCGCCUCUAAUU-UUGUUUUGCCACAAACUUUUGCCGGUUUCCCCGACAGCCGUGGCA ((..(((((((((((((...........)))))))))))))..))...................-......((((((......(((.(((.....))).)))..)))))) ( -20.60) >DroSim_CAF1 12269 110 + 1 GAACAGAAUUUAAUAAAACCUAUUUCCAUUUAUUAAAUUCUUCUCUUCAUCGUCGCCUCUAAUUUUUGUUUUGCCACAAACUUUUGCCGGUUUCCCUGACAGCCGUGGCA ((..(((((((((((((...........)))))))))))))..)).........(((.......(((((......))))).......(((((........))))).))). ( -19.90) >DroEre_CAF1 12246 108 + 1 GAACAGAAUUUAAUAAAACCUAUUUCCAUUUAUUAAAUUCUUCUGCUCAUCGUCGCCUCUAAUU-UUGUUUUGCCACAAACUUUGGCCGCUUUU-CCGGCAGCCGUGGCA (((((((((((((((((...........))))))))))))....((........))........-.)))))((((((.......(((.(((...-..))).))))))))) ( -24.21) >DroYak_CAF1 17208 109 + 1 GAACAGAAUUUAAUAAAACCUAUUUCCAUUUAUUAAAUUCUUCUCUUCAUCGUCGCCUCUAAUU-UUGUUUUGCCACAAACUUUUGCCGCCUUUCCCGACAGCCGUGGCA ((..(((((((((((((...........)))))))))))))..))...................-......((((((...((.(((..........))).))..)))))) ( -17.20) >consensus GAACAGAAUUUAAUAAAACCUAUUUCCAUUUAUUAAAUUCUUCUCUUCAUCGUCGCCUCUAAUU_UUGUUUUGCCACAAACUUUUGCCGGUUUUCCCGACAGCCGUGGCA ((..(((((((((((((...........)))))))))))))..))..........................((((((...(....)..((((........)))))))))) (-16.44 = -16.88 + 0.44)

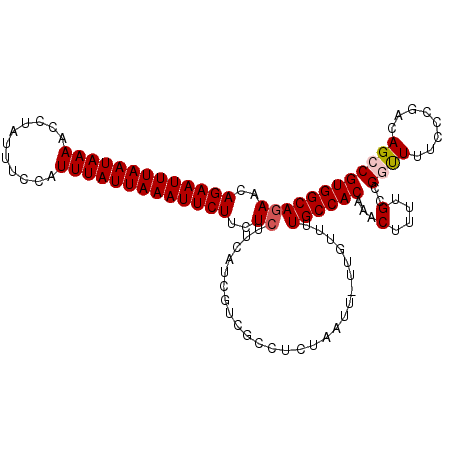

| Location | 9,110,033 – 9,110,142 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110033 109 - 22407834 UGCCACGGCUGUCGGGAAAACCGGCCAAAGUUUGUGGCAAAGCAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAAUAAAUGGAAAUAGGUUUUAUUAAAUUCUGUUC (((((((((((((((.....)))))...)).))))))))......-..(((..(....)..))).(((.(((((((((((((...........))))))))))))).))) ( -31.00) >DroSec_CAF1 12149 109 - 1 UGCCACGGCUGUCGGGGAAACCGGCAAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAAUAAAUGGAAGAAGGUUUUAUUAAAUUCUGUUC (((((((((((((((.....)))))...)).))))))))......-..(((..(....)..))).(((.(((((((((((((...........))))))))))))).))) ( -31.00) >DroSim_CAF1 12269 110 - 1 UGCCACGGCUGUCAGGGAAACCGGCAAAAGUUUGUGGCAAAACAAAAAUUAGAGGCGACGAUGAAGAGAAGAAUUUAAUAAAUGGAAAUAGGUUUUAUUAAAUUCUGUUC (((((((((((((.((....)))))...)).)))))))).........(((..(....)..))).(((.(((((((((((((...........))))))))))))).))) ( -30.20) >DroEre_CAF1 12246 108 - 1 UGCCACGGCUGCCGG-AAAAGCGGCCAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAGCAGAAGAAUUUAAUAAAUGGAAAUAGGUUUUAUUAAAUUCUGUUC .(((..((((((...-....))))))....(((((......))))-)......)))......(((((((...((((((((((...........))))))))))))))))) ( -30.70) >DroYak_CAF1 17208 109 - 1 UGCCACGGCUGUCGGGAAAGGCGGCAAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAAUAAAUGGAAAUAGGUUUUAUUAAAUUCUGUUC (((((((((((((......)))))).......)))))))......-..(((..(....)..))).(((.(((((((((((((...........))))))))))))).))) ( -29.51) >consensus UGCCACGGCUGUCGGGAAAACCGGCAAAAGUUUGUGGCAAAACAA_AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAAUAAAUGGAAAUAGGUUUUAUUAAAUUCUGUUC (((((((((((((((.....)))))...)).)))))))).(((.....(((..(....)..))).....(((((((((((((...........)))))))))))))))). (-26.94 = -27.22 + 0.28)

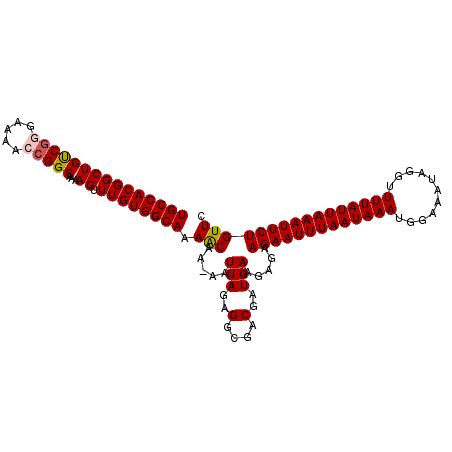

| Location | 9,110,065 – 9,110,182 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.86 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110065 117 - 22407834 UGAAUUGCGCCAGUAGUCGUGGAUGCGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAACCGGCCAAAGUUUGUGGCAAAGCAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAA ...((((((((..((((..((....(....).......(((((((((((((((((.....)))))...)).)))))))))).)).-.))))..)))).))))................ ( -34.80) >DroSec_CAF1 12181 111 - 1 UGAAUUGCGCCAGUAU------AUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGGAAACCGGCAAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAA ...((((((((.(((.------...)))..........(((((((((((((((((.....)))))...)).))))))))))....-.......)))).))))................ ( -31.70) >DroSim_CAF1 12301 118 - 1 UGAAUUGCGCCAGUAAUCGUGGAUGCCCAAGAUAAACAUUUGCCACGGCUGUCAGGGAAACCGGCAAAAGUUUGUGGCAAAACAAAAAUUAGAGGCGACGAUGAAGAGAAGAAUUUAA ...((((((((....(((.(((....))).))).....(((((((((((((((.((....)))))...)).))))))))))............)))).))))................ ( -34.70) >DroEre_CAF1 12278 116 - 1 UGAAUUGCGCCAGUAGUCGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGCCGG-AAAAGCGGCCAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAGCAGAAGAAUUUAA ((.((((((((.((((((((((..(((.........)))...))))))))))...-.......((((.......)))).......-.......)))).))))...))........... ( -33.50) >DroYak_CAF1 17240 117 - 1 UGAAUUGCGCCAGUGGUUGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAGGCGGCAAAAGUUUGUGGCAAAACAA-AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAA ...((((((((..(((((.(((((((.........)))))(((((((((((((......)))))).......)))))))...)).-)))))..)))).))))................ ( -32.41) >consensus UGAAUUGCGCCAGUAGUCGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAACCGGCAAAAGUUUGUGGCAAAACAA_AAUUAGAGGCGACGAUGAAGAGAAGAAUUUAA ...((((((((....(((.((......)).))).....(((((((((((((((((.....)))))...)).))))))))))............)))).))))................ (-26.66 = -27.86 + 1.20)

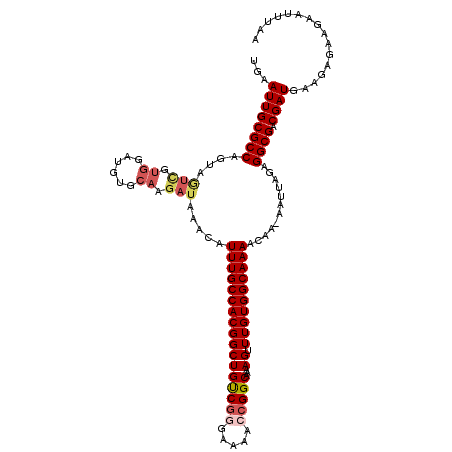
| Location | 9,110,102 – 9,110,220 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -29.48 |
| Energy contribution | -31.60 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9110102 118 - 22407834 -CCAACUACAGAUAUUUGCAUUUCUUGAGGU-UUGCUUUAUGAAUUGCGCCAGUAGUCGUGGAUGCGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAACCGGCCAAAGUUUGUGGCAA -....((((((((....(((.(((.(((((.-...))))).))).)))(((.((((((((((...(....)...........))))))))).)((.....)))))....))))))))... ( -38.24) >DroSec_CAF1 12218 112 - 1 -CCAACUACAGAUAUUUGCAUUUCUUGAGGU-UUGCUUUAUGAAUUGCGCCAGUAU------AUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGGAAACCGGCAAAAGUUUGUGGCAA -................(((.(((.(((((.-...))))).))).)))(((.(((.------...)))...((((((.((((((.((.....))((....)))))))).))))))))).. ( -34.50) >DroSim_CAF1 12339 119 - 1 -CCAACUACAGAUAUUUGCAUUUCUUGAGGUUUUGCUUUAUGAAUUGCGCCAGUAAUCGUGGAUGCCCAAGAUAAACAUUUGCCACGGCUGUCAGGGAAACCGGCAAAAGUUUGUGGCAA -....((((((((.(((((...(((((.(((.....(((((((.((((....))))))))))).)))))))).........((....)).....((....)).))))).))))))))... ( -35.50) >DroEre_CAF1 12315 117 - 1 -CCAACUACAGAUAUUUGCAUUUCUUGAGGU-UUGCUUUAUGAAUUGCGCCAGUAGUCGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGCCGG-AAAAGCGGCCAAAGUUUGUGGCAA -....((((((((....(((.(((.(((((.-...))))).))).))).((.((((((((((..(((.........)))...)))))))))).))-.............))))))))... ( -38.10) >DroYak_CAF1 17277 119 - 1 ACCAACUACAGAUAUUUGCAUUUCUUGAGGU-UUGCUUUAUGAAUUGCGCCAGUGGUUGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAGGCGGCAAAAGUUUGUGGCAA .....((((((((.((((((.(((.(((((.-...))))).))).)))((((((.((.(..(((((.........)))))..).)).)))(((......))))))))).))))))))... ( -38.80) >consensus _CCAACUACAGAUAUUUGCAUUUCUUGAGGU_UUGCUUUAUGAAUUGCGCCAGUAGUCGUGGAUGUGCAAGAUAAACAUUUGCCACGGCUGUCGGGAAAACCGGCAAAAGUUUGUGGCAA .....((((((((....(((.(((.(((((.....))))).))).)))(((.((((((((((((((.........))))...))))))))).).((....)))))....))))))))... (-29.48 = -31.60 + 2.12)

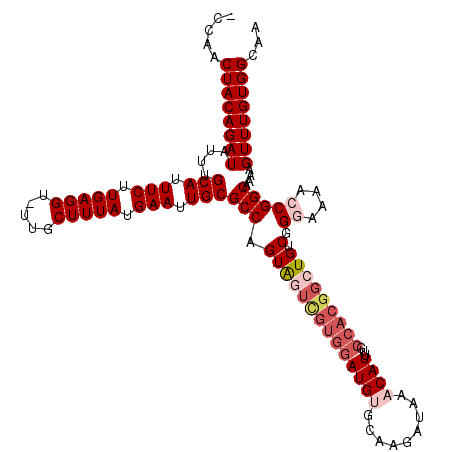
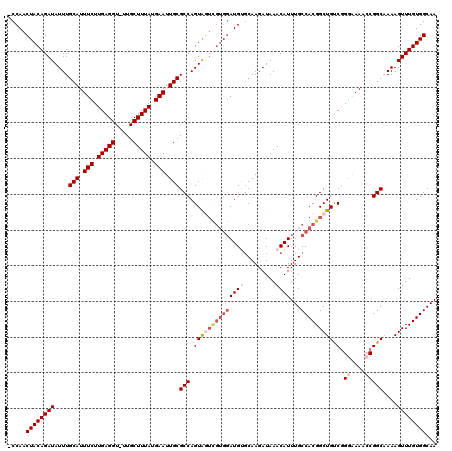
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:34 2006