
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 910,762 – 910,861 |
| Length | 99 |
| Max. P | 0.672470 |

| Location | 910,762 – 910,861 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 910762 99 + 22407834 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACAGC--U---GAA---------AGAGUCGGGGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..((((((((.......((((.....))))......))--)---)))---------))......(((.........))). ( -27.52) >DroPse_CAF1 38605 105 + 1 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACGCG--A---GACACCCCAC--AAAG-GGAAGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..(((((((((....)))))))))..........(((.--.---....(((...--...)-))..)))............ ( -27.20) >DroEre_CAF1 46855 98 + 1 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACAGC--U---GAA---------AGAGUCG-GGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..((((((((.......((((.....))))......))--)---)))---------)).....-(((.........))). ( -27.52) >DroYak_CAF1 41248 108 + 1 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACAGC--U---GAAAGAGCUGAAAGAGUCGGGGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..(((((((((....)))))))))..........((((--(---.....)))))..........(((.........))). ( -31.40) >DroAna_CAF1 39146 106 + 1 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGGCGGGGCCGGGGACAC-------ACCAGCGCAGCAAGACAAAAGCCA ((((((((...(((.(((..((((.....))))..(((((((((....)))))))))........)))...)))(....)..-------..)))).))))............. ( -35.40) >DroPer_CAF1 38367 105 + 1 GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACGCG--A---GACACCCCAC--AAAG-GGAAGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..(((((((((....)))))))))..........(((.--.---....(((...--...)-))..)))............ ( -27.20) >consensus GUUGGCUGAAAGGCAGCCAAGUGUUCAAUGCACAACUUUUAGCGCAAUUGUUAAAAGAAUUAAAAGACAGC__U___GAAAC_______AGAGUCGAAGCGAAACAAAAGCCA .(((((((.....)))))))((((.....))))..(((((((((....)))))))))........................................................ (-20.80 = -20.80 + 0.00)

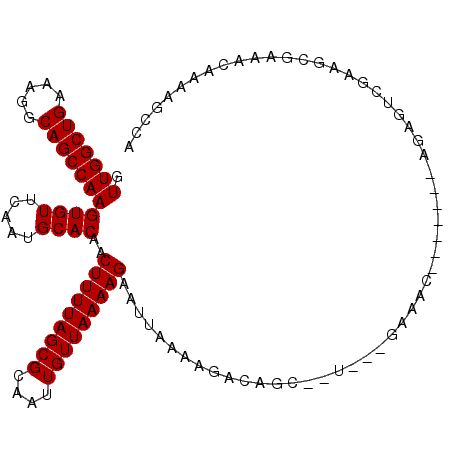
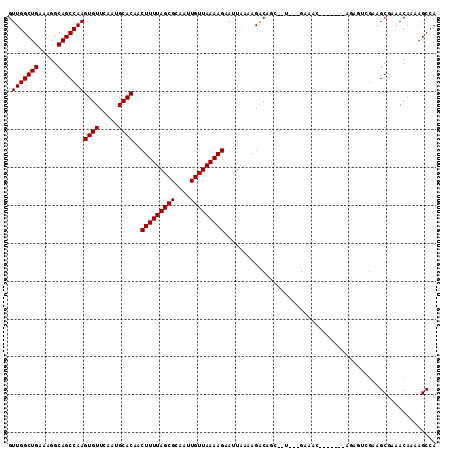
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:57 2006