| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,108,365 – 9,108,509 |
| Length | 144 |
| Max. P | 0.991456 |

| Location | 9,108,365 – 9,108,475 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
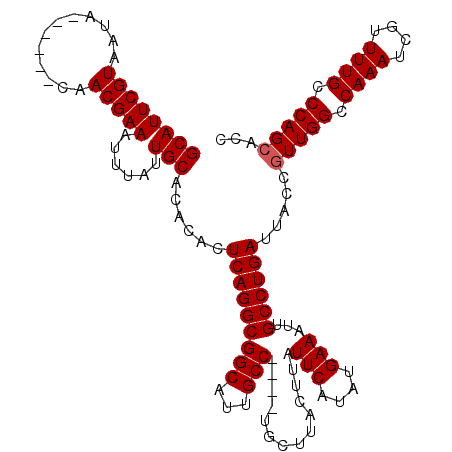
>2L_DroMel_CAF1 9108365 110 + 22407834 GCAUUCGUAAUA------CAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCC----UGCUUACUUAUUCAUAUGAAAUUGCCUGAUUACCGUUGGCCAAAUCGUUUUGCCCAGCACC ((((((((....------..)))))......)))......(((((((((...)))----..........(((....)))...)))))).....(((((.((((....)))).)))))... ( -27.50) >DroSec_CAF1 10520 114 + 1 GCAUUCGUAAUA------CAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCCUGCCUGCUUACUUAUUCAUAUGAAAAUGCCUGAUUACCGUUGGCCAAAUCGUUUUGCCCAGCACC ((((((((....------..)))))......)))......((((((((((.....))))..........(((....)))...)))))).....(((((.((((....)))).)))))... ( -30.00) >DroSim_CAF1 10603 114 + 1 GCAUUCGUAAUA------CAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCCUGCCUGCUUACUUAUUCAUAUGAAAUUGCCUGAUUACCGUUGGCCAAAUCGUUUUGCCCAGAACC ((((((((....------..)))))......)))......((((((((((.....))))..........(((....)))...))))))......((((.((((....)))).)))).... ( -26.00) >DroEre_CAF1 10713 116 + 1 GCAUUCGUAAUACUCGAACAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCC----UGCUUACUUAUUCAUAUGAAAUUGCCUGAUUACCGUUGGCCAAAUCGUUUUGCCCAGCACC ......((((((.(((.....)))....))))))......(((((((((...)))----..........(((....)))...)))))).....(((((.((((....)))).)))))... ( -27.10) >DroYak_CAF1 15549 110 + 1 GCAUUCGUAAUA------CAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCC----UGCUUACUUAUUCAUAUGAAAUUGCCUGAUUACCGUUGGCCAAAUCGCUUUGCCCAGCGCC ((((((((....------..)))))......)))......(((((((((...)))----..........(((....)))...))))))....((((((.((((....)))).)))))).. ( -27.80) >consensus GCAUUCGUAAUA______CAACGAAAUUUAUUGCACACACUCAGGCGGCAUUGCC____UGCUUACUUAUUCAUAUGAAAUUGCCUGAUUACCGUUGGCCAAAUCGUUUUGCCCAGCACC ((((((((............)))))......)))......(((((((((...)))..............(((....)))...)))))).....(((((.((((....)))).)))))... (-24.12 = -24.32 + 0.20)


| Location | 9,108,399 – 9,108,509 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.68 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -25.13 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
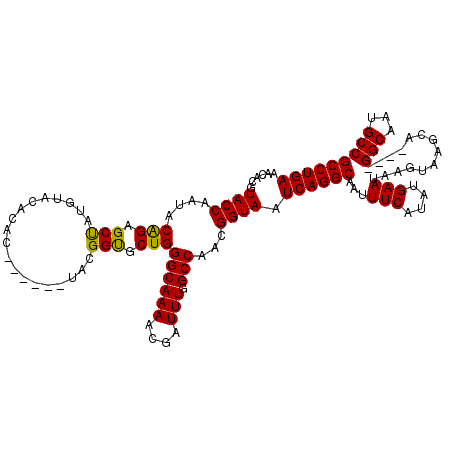
>2L_DroMel_CAF1 9108399 110 - 22407834 AACACGUACCAAUACAGAGCUAUGUACACAC------UACGGUGCUGGGCAAAACGAUUUGGCCAACGGUAAUCAGGCAAUUUCAUAUGAAUAAGUAAGCA----GGCAAUGCCGCCUGA ......((((..((((......))))..(((------....))).(((.((((....)))).)))..)))).((((((...(((....)))..........----(((...))))))))) ( -29.50) >DroSec_CAF1 10554 114 - 1 AACACGUACCAAUACAGAGCUAUAUACACAC------UACGGUGCUGGGCAAAACGAUUUGGCCAACGGUAAUCAGGCAUUUUCAUAUGAAUAAGUAAGCAGGCAGGCAAUGCCGCCUGA ......((((..................(((------....))).(((.((((....)))).)))..)))).((((((...(((....)))..........((((.....)))))))))) ( -31.60) >DroSim_CAF1 10637 114 - 1 AACACGUACCAAUACAGAGCUAUGUACACAC------UAUGGUUCUGGGCAAAACGAUUUGGCCAACGGUAAUCAGGCAAUUUCAUAUGAAUAAGUAAGCAGGCAGGCAAUGCCGCCUGA ......((((....((((((((((.......------))))))))))(((...........)))...)))).((((((...(((....)))..........((((.....)))))))))) ( -35.50) >DroEre_CAF1 10753 116 - 1 AACACGUACCAAUACGGAGCCGUGUACACACGAAGCCCACGGUGCUGGGCAAAACGAUUUGGCCAACGGUAAUCAGGCAAUUUCAUAUGAAUAAGUAAGCA----GGCAAUGCCGCCUGA .((.(((.((((..((....((((....))))..(((((......)))))....))..))))...)))))..((((((...(((....)))..........----(((...))))))))) ( -33.70) >DroYak_CAF1 15583 116 - 1 AACACGUACCAAUACAGAGCUAUGUACAUACGACGACGACGGCGCUGGGCAAAGCGAUUUGGCCAACGGUAAUCAGGCAAUUUCAUAUGAAUAAGUAAGCA----GGCAAUGCCGCCUGA ......((((....(((.(((.(((.(.......))))..))).)))((((((....))).)))...)))).((((((...(((....)))..........----(((...))))))))) ( -27.60) >consensus AACACGUACCAAUACAGAGCUAUGUACACAC______UACGGUGCUGGGCAAAACGAUUUGGCCAACGGUAAUCAGGCAAUUUCAUAUGAAUAAGUAAGCA____GGCAAUGCCGCCUGA ......((((....(((.(((...................))).)))((((((....))).)))...)))).((((((...(((....)))..............(((...))))))))) (-25.13 = -24.65 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:28 2006