
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
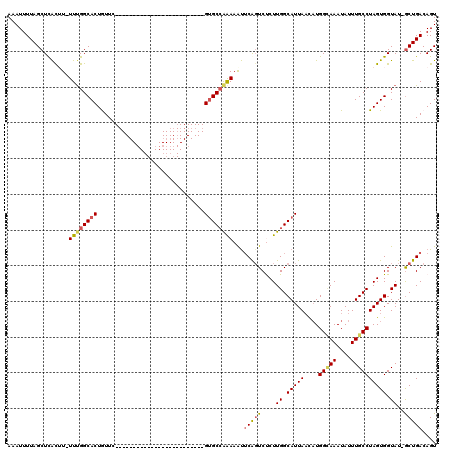
| Location | 9,107,017 – 9,107,147 |
| Length | 130 |
| Max. P | 0.969242 |

| Location | 9,107,017 – 9,107,111 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9107017 94 + 22407834 AAAUUUUAGCUCAUUU-UUUGGCACUAUUC-------------------------GUGCCAAAAAUUCAGUCUCUUGGCAUUAACAUGGUAAAUAUUUGCCUAGUGGUAUUACUGACAGU ..............((-((((((((.....-------------------------)))))))))).(((((....((.(((((....(((((....)))))))))).))..))))).... ( -23.80) >DroSec_CAF1 9191 93 + 1 AAAUUUUAGCUCACGU-UUUGGCACUGUUU-------------------------GUGCCAAAAAUUCAGUCUCUUGGCAUUAACAUGGCAAAUAUUUGCCUAGUGGUAU-GCUGACAGU ........(((....(-((((((((.....-------------------------)))))))))....)))((.(..((((...((((((((....)))))..)))..))-))..).)). ( -28.70) >DroSim_CAF1 9263 93 + 1 AAAUUUUAGCUUACGU-UUCGGCACUGUUC-------------------------GUGCCAAAAAUGCAGUCUCUUGGCAUUAACAUGGCAAAUAUUUGCCUAGUGGUAU-GCUGACAGU .....(((((.(((..-...(((((.....-------------------------)))))...(((((.........)))))..((((((((....)))))..)))))).-))))).... ( -25.30) >DroEre_CAF1 9354 93 + 1 AAAUUUUAGCUCGCUU-UUUGGCACUAUUC-------------------------GUGCCAGAAGCUCGGUCCCUUGGCAUUAUUCUGGUAAAUAUUUGCCUAGUGGUAU-GCUGACAGU .....(((((..((((-((.(((((.....-------------------------))))))))))).....((((.((((.((((......))))..)))).)).))...-))))).... ( -28.20) >DroYak_CAF1 13996 119 + 1 AAAUUUUAGCUCACUUUUUUGGCACUGUUCUGUUCUGUGGAAAAAAGCGGAGGAAGAGCGGAAAAUUCGUUCCCUUGGCAUUAAUAUGGGAAAUAUUUGCCUAGUGGUAU-GCUGACAGU .....((((((((((.....(((.(((((((.((((((........))))))..))))))).((((...(((((.............)))))..))))))).)))))...-))))).... ( -33.92) >consensus AAAUUUUAGCUCACUU_UUUGGCACUGUUC_________________________GUGCCAAAAAUUCAGUCUCUUGGCAUUAACAUGGCAAAUAUUUGCCUAGUGGUAU_GCUGACAGU .................((((((((..............................))))))))...(((((....((.(((((....(((((....)))))))))).))..))))).... (-15.87 = -16.23 + 0.36)


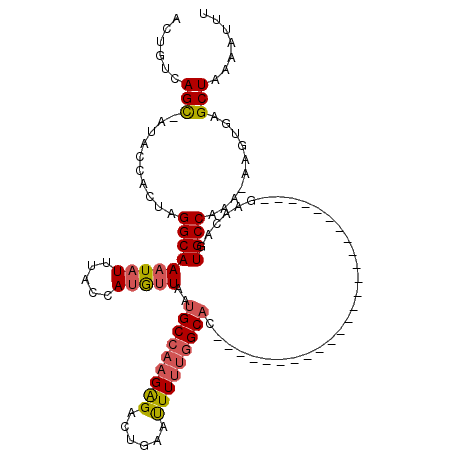
| Location | 9,107,017 – 9,107,111 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.99 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9107017 94 - 22407834 ACUGUCAGUAAUACCACUAGGCAAAUAUUUACCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC-------------------------GAAUAGUGCCAAA-AAAUGAGCUAAAAUUU ....(((((.......((.(((((((((.....)))))..)))).))..))))).((((((((((-------------------------.....))))))))-)).............. ( -23.60) >DroSec_CAF1 9191 93 - 1 ACUGUCAGC-AUACCACUAGGCAAAUAUUUGCCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC-------------------------AAACAGUGCCAAA-ACGUGAGCUAAAAUUU .((.(..((-((...((..(((((....)))))..))..))))..).)).((.(..(((((((((-------------------------.....))))))))-)..).))......... ( -23.60) >DroSim_CAF1 9263 93 - 1 ACUGUCAGC-AUACCACUAGGCAAAUAUUUGCCAUGUUAAUGCCAAGAGACUGCAUUUUUGGCAC-------------------------GAACAGUGCCGAA-ACGUAAGCUAAAAUUU ....((.((-((.......(((((....))))).((((..(((((((((......))))))))).-------------------------.)))))))).)).-................ ( -24.60) >DroEre_CAF1 9354 93 - 1 ACUGUCAGC-AUACCACUAGGCAAAUAUUUACCAGAAUAAUGCCAAGGGACCGAGCUUCUGGCAC-------------------------GAAUAGUGCCAAA-AAGCGAGCUAAAAUUU ......(((-...((.((.((((..(((((....))))).)))).)))).....((((.((((((-------------------------.....))))))..-))))..)))....... ( -27.30) >DroYak_CAF1 13996 119 - 1 ACUGUCAGC-AUACCACUAGGCAAAUAUUUCCCAUAUUAAUGCCAAGGGAACGAAUUUUCCGCUCUUCCUCCGCUUUUUUCCACAGAACAGAACAGUGCCAAAAAAGUGAGCUAAAAUUU ......(((-......((.(((((((((.....)))))..)))).))((((......))))..........(((((((((.(((...........)))..))))))))).)))....... ( -20.50) >consensus ACUGUCAGC_AUACCACUAGGCAAAUAUUUACCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC_________________________GAACAGUGCCAAA_AAGUGAGCUAAAAUUU ......(((..........(((((((((.....)))))..(((((((((......)))))))))................................))))..........)))....... (-12.59 = -12.99 + 0.40)

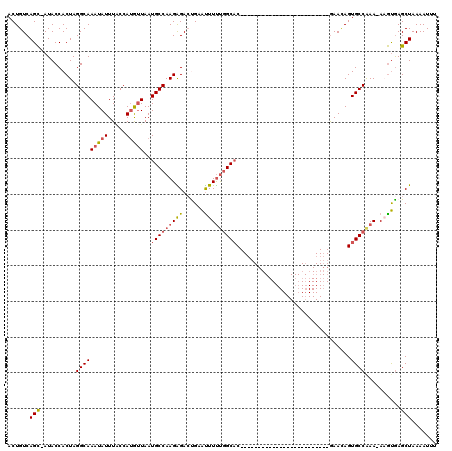
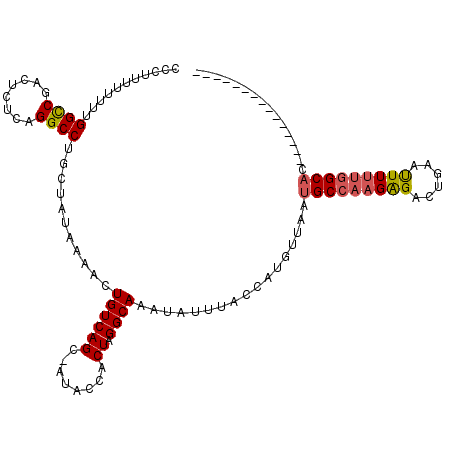
| Location | 9,107,046 – 9,107,147 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9107046 101 - 22407834 CCUUUUUUUUUGGCCGAUUCUCAGGCCUGCUAUAAAACUGUCAGUAAUACCACUAGGCAAAUAUUUACCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC--------------- ...........((((........))))(((((.((((...(((((.......((.(((((((((.....)))))..)))).))..))))).))))))))).--------------- ( -22.50) >DroSec_CAF1 9220 99 - 1 CCCUUU-UUUUGGUCGACUCUCAGGCCAUCUAUAAAACUGUCAGC-AUACCACUAGGCAAAUAUUUGCCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC--------------- ......-...(((((........))))).................-.........(((((....))))).......(((((((((......))))))))).--------------- ( -21.60) >DroSim_CAF1 9292 99 - 1 CCCUUU-UUUUGGCCGACUCUCAGGCCUGCUAUAAAACUGUCAGC-AUACCACUAGGCAAAUAUUUGCCAUGUUAAUGCCAAGAGACUGCAUUUUUGGCAC--------------- ......-....((((........))))(((((((....))).)))-)........(((((....))))).......(((((((((......))))))))).--------------- ( -25.70) >DroEre_CAF1 9383 100 - 1 CCCUUUUUUUUGGCCGAUUCUCAGGCCUGCUAUAAAACUGUCAGC-AUACCACUAGGCAAAUAUUUACCAGAAUAAUGCCAAGGGACCGAGCUUCUGGCAC--------------- ............(((((..(((.(((((((((((....))).)))-).....((.((((..(((((....))))).)))).)))).)))))..)).)))..--------------- ( -24.30) >DroYak_CAF1 14036 115 - 1 CCCGUUUUUUUGGCCGACUCUCAGGCCUGCUAUAAAACUGUCAGC-AUACCACUAGGCAAAUAUUUCCCAUAUUAAUGCCAAGGGAACGAAUUUUCCGCUCUUCCUCCGCUUUUUU ..(((((((((((((........))))(((((((....))).)))-)........(((((((((.....)))))..)))))))))))))........................... ( -22.40) >consensus CCCUUUUUUUUGGCCGACUCUCAGGCCUGCUAUAAAACUGUCAGC_AUACCACUAGGCAAAUAUUUACCAUGUUAAUGCCAAGAGACUGAAUUUUUGGCAC_______________ ...........((((........))))...........((((((........)).)))).................(((((((((......)))))))))................ (-16.80 = -17.04 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:21 2006