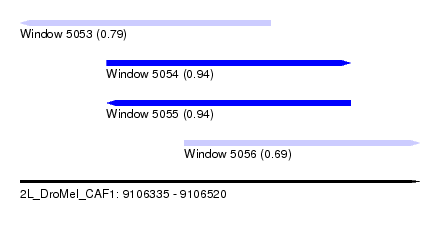
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,106,335 – 9,106,520 |
| Length | 185 |
| Max. P | 0.943508 |

| Location | 9,106,335 – 9,106,451 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -15.20 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
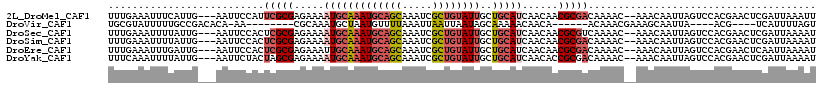
>2L_DroMel_CAF1 9106335 116 - 22407834 CGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAAUGGAAUU---CAAUGAAAUUUCAAAACUUACCCAUUAUUGAUAAAUAUUUAAUGCGCAUAAUUUA .(((((((.((((((((((.........)))))))))).))).........(((((....(---((((((..................)))))))....)))))...))))........ ( -26.77) >DroVir_CAF1 39634 103 - 1 -----UGUUGUUUUUGCUAUUAAUUAAUUUAAAACAUUAGCAUUUGCG--------UU-UGUGUCGGCAAAAAUACGCAAAUUGC--GUGGCUAUUUAAUAUUUUAUGCAUAUAAUUUA -----(((((((((((((.((((.....)))).(((...((....)).--------..-)))...)))))))))).)))...(((--((((............)))))))......... ( -17.50) >DroSec_CAF1 8510 116 - 1 CGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU---CAAUAAAAUUUCAAAACUUACCCAUUAUUGACAAAUAUUUAAUGCGCAUAAUUUA .(((((((.((((((((((.........)))))))))).))).((((.(....).)))).(---((((((..................)))))))............))))........ ( -26.97) >DroSim_CAF1 8593 116 - 1 CGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU---CAAUAAAAUUUCAAAACUUACCCAUUAUUGAUAAAUAUUUAAUGCGCAUAAUUUA .(((((((.((((((((((.........)))))))))).))).((((.(....).)))).(---((((((..................)))))))............))))........ ( -27.07) >DroEre_CAF1 8685 116 - 1 CGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAAUUUCUCGCGAGUGGAAUU---CAAUCAAAUUUCAAAACUUGCCCAUGUUUGAUAAAUAUUUAAUGCGCAUAAUUUA .(((((((.((((((((((.........)))))))))).))).......((((((((((((---......))))))...))))))......................))))........ ( -30.40) >DroYak_CAF1 13306 116 - 1 CGCGGUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCUAGUAGAAUU---CAAUAAAAUUUGAAAACUGGCCCAUGAUUGAUAAAUAUUUAAUGAGCAUAAUUUA .(((((((.((((((((((.........)))))))))).))).......((((((....((---(((......))))).)))))))).(.(((((.......))))).)))........ ( -30.40) >consensus CGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU___CAAUAAAAUUUCAAAACUUACCCAUGAUUGAUAAAUAUUUAAUGCGCAUAAUUUA .....(((.((((((((((.........)))))))))).))).......(((....((((....((((((..................))))))......))))....)))........ (-15.20 = -16.07 + 0.87)

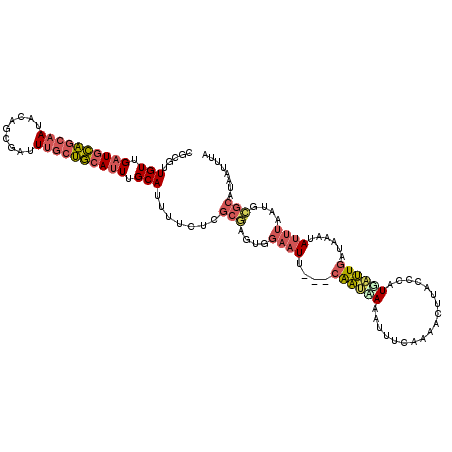
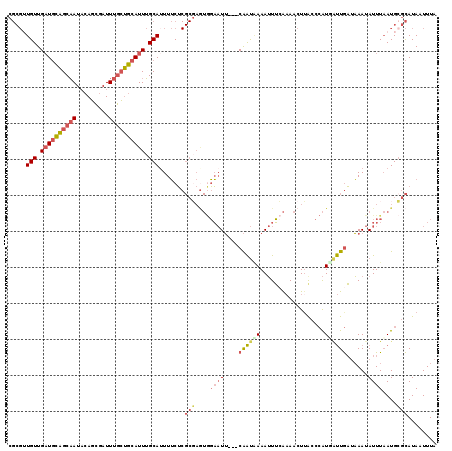
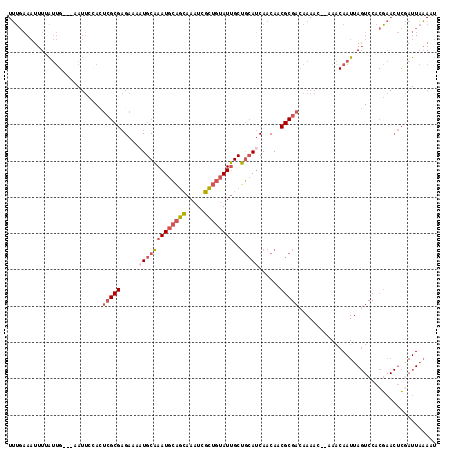
| Location | 9,106,375 – 9,106,488 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -12.74 |
| Energy contribution | -13.78 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9106375 113 + 22407834 UUUGAAAUUUCAUUG---AAUUCCAUUCGCGAGAAAAUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAUU .....(((((.((((---(.(((...(((((.....(((((((((((((.....))))))))..)))))......)))))......--.........(....)))).))))).))))) ( -25.30) >DroVir_CAF1 39672 95 + 1 UGCGUAUUUUUGCCGACACA-AA--------CGCAAAUGCUAAUGUUUUAAAUUAAUUAAUAGCAAAAACAACA------ACAAACGAAAGCAAUUA----ACG----UCAUUUUAGU .(((......))).(((...-..--------......(((((..(((.......)))...))))).........------.....(....)......----..)----))........ ( -9.50) >DroSec_CAF1 8550 113 + 1 UUUGAAAUUUUAUUG---AAUUCCACUCGCGAGAAAAUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGUCAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAU ......(((((((((---(.(((....((((.....(((((((((((((.....))))))))..)))))......)))).......--.........(....)))).)))).)))))) ( -22.60) >DroSim_CAF1 8633 113 + 1 UUUGAAAUUUUAUUG---AAUUCCACUCGCGAGAAAAUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAU ......(((((((((---(.(((...(((((.....(((((((((((((.....))))))))..)))))......)))))......--.........(....)))).)))).)))))) ( -24.40) >DroEre_CAF1 8725 113 + 1 UUUGAAAUUUGAUUG---AAUUCCACUCGCGAGAAAUUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCAAUUAAAAU .......((((((((---(.(((...(((((......((((((((((((.....))))))))..)))).......)))))......--.........(....)))).))))))))).. ( -27.72) >DroYak_CAF1 13346 113 + 1 UUUCAAAUUUUAUUG---AAUUCUACUAGCGAGAAAAUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACACCGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAU ......(((((((((---(.(((.....(((.....(((((((((((((.....))))))))..)))))......)))(((.....--.........)))...))).)))).)))))) ( -23.94) >consensus UUUGAAAUUUUAUUG___AAUUCCACUCGCGAGAAAAUGCAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC__AAACAAUUAGUCCACGAACUCGAUUAAAAU ..........................(((((.....(((((((((((((.....))))))))..)))))......)))))...................................... (-12.74 = -13.78 + 1.04)

| Location | 9,106,375 – 9,106,488 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -17.21 |
| Energy contribution | -18.55 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
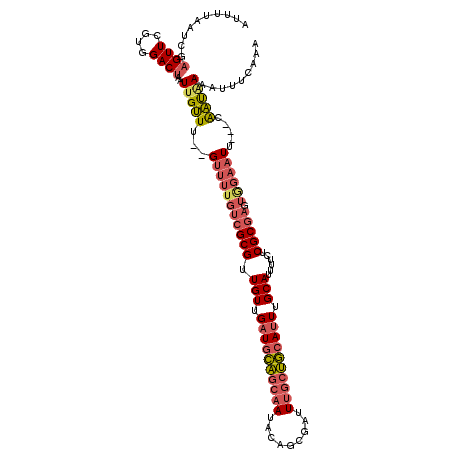
>2L_DroMel_CAF1 9106375 113 - 22407834 AAUUUAAUCGAGUUCGUGGACUAAUUGUUU--GUUUUGUCGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAAUGGAAUU---CAAUGAAAUUUCAAA .........(((((((..(((.....))).--.)....(((((.(((.((((((((((.........)))))))))).)))......)))))...)))))---).............. ( -32.30) >DroVir_CAF1 39672 95 - 1 ACUAAAAUGA----CGU----UAAUUGCUUUCGUUUGU------UGUUGUUUUUGCUAUUAAUUAAUUUAAAACAUUAGCAUUUGCG--------UU-UGUGUCGGCAAAAAUACGCA ....((((((----.((----.....))..))))))..------(((((((((((((.((((.....)))).(((...((....)).--------..-)))...)))))))))).))) ( -15.50) >DroSec_CAF1 8550 113 - 1 AUUUUAAUCGAGUUCGUGGACUAAUUGUUU--GUUUUGACGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU---CAAUAAAAUUUCAAA .........(((((((..(((.....))).--.).....((((.(((.((((((((((.........)))))))))).)))......))))....)))))---).............. ( -31.00) >DroSim_CAF1 8633 113 - 1 AUUUUAAUCGAGUUCGUGGACUAAUUGUUU--GUUUUGUCGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU---CAAUAAAAUUUCAAA .........(((((((..(((.....))).--.)....(((((.(((.((((((((((.........)))))))))).)))......)))))...)))))---).............. ( -32.30) >DroEre_CAF1 8725 113 - 1 AUUUUAAUUGAGUUCGUGGACUAAUUGUUU--GUUUUGUCGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAAUUUCUCGCGAGUGGAAUU---CAAUCAAAUUUCAAA ..(((.((((((((((..(((.....))).--.)....(((((((((.((((((((((.........)))))))))).)))).....)))))...)))))---)))).)))....... ( -35.20) >DroYak_CAF1 13346 113 - 1 AUUUUAAUCGAGUUCGUGGACUAAUUGUUU--GUUUUGUCGCGGUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCUAGUAGAAUU---CAAUAAAAUUUGAAA .(((((..((((..(((((...(((.....--)))...)))))((((.((((((((((.........)))))))))).))))...))))....)))))((---(((......))))). ( -30.10) >consensus AUUUUAAUCGAGUUCGUGGACUAAUUGUUU__GUUUUGUCGCGUUGUUGAUGCAGCAAUACAGCGAUUUGCUGCAUUUGCAUUUUCUCGCGAGUGGAAUU___CAAUAAAAUUUCAAA ..........((((....))))..(((((...(((((((((((.(((.((((((((((.........)))))))))).)))......))))).)))))).....)))))......... (-17.21 = -18.55 + 1.34)

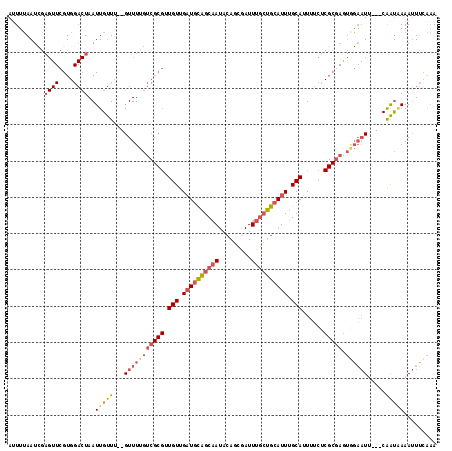
| Location | 9,106,411 – 9,106,520 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -17.11 |
| Consensus MFE | -10.50 |
| Energy contribution | -11.83 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9106411 109 + 22407834 CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAUUUAAUUUAUAAUCAGUUUGAUGAAUAAA--AAAAU ....((((((((.........))))))))(((.....(.(((.....--.........)))).((((((.(((((...........))))))))))).))).....--..... ( -19.44) >DroVir_CAF1 39702 99 + 1 CUAAUGUUUUAAAUUAAUUAAUAGCAAAAACAACA------ACAAACGAAAGCAAUUA----ACG----UCAUUUUAGUCUAUUGACUAUCAGCUCAAAUAAUACAACAAAGG ......................(((..........------.....(....)......----...----......(((((....)))))...))).................. ( -7.80) >DroSec_CAF1 8586 109 + 1 CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGUCAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAUUAAUUUAUAAUCAGUUUGAUGAAUAAA--AAAAU ...(((((((((.........))))))))).................--............((((((((.(((((.((.....)).))))))))))).))......--..... ( -19.50) >DroSim_CAF1 8669 109 + 1 CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAUUAAUUUAUAAUCAGUUUGAUGAAUAAA--AAAAU ....((((((((.........))))))))(((.....(.(((.....--.........)))).((((((.(((((.((.....)).))))))))))).))).....--..... ( -20.44) >DroEre_CAF1 8761 110 + 1 CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC--AAACAAUUAGUCCACGAACUCAAUUAAAAUUAAUUUAUAAUCAGUUUGAUGAAUAAAA-AAAAU ....((((((((.........))))))))(((.....(.(((.....--.........)))).((((((..((((.((.....)).)))).)))))).)))......-..... ( -16.04) >DroYak_CAF1 13382 111 + 1 CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACACCGCGACAAAAC--AAACAAUUAGUCCACGAACUCGAUUAAAAUUAAUAUAUAAUCAGUUUGAUGAAUAAAAAAAAAU ....((((((((.........))))))))(((.....(.(((.....--.........)))).((((((.(((((...........))))))))))).)))............ ( -19.44) >consensus CAAAUGCAGCAAAUCGCUGUAUUGCUGCAUCAACAACGCGACAAAAC__AAACAAUUAGUCCACGAACUCGAUUAAAAUUAAUUUAUAAUCAGUUUGAUGAAUAAA__AAAAU ...(((((((((.........))))))))).................................((((((.(((((...........)))))))))))................ (-10.50 = -11.83 + 1.34)
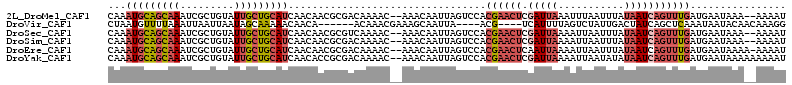
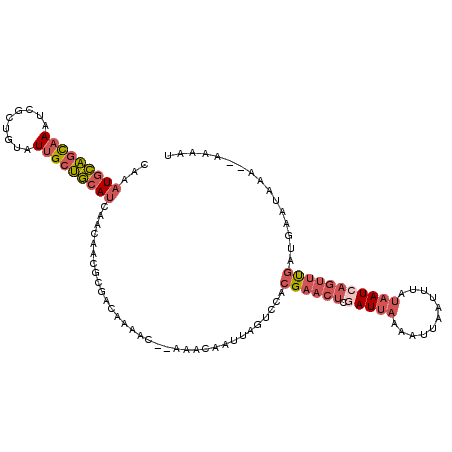
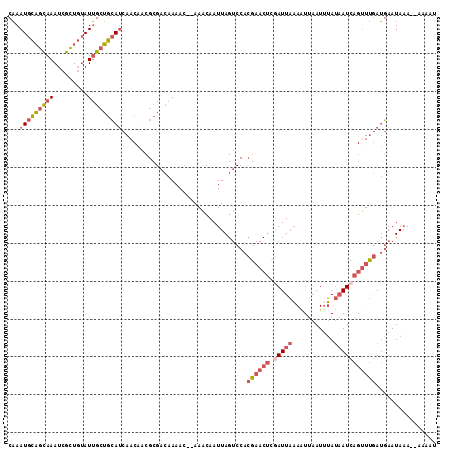
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:19 2006