| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
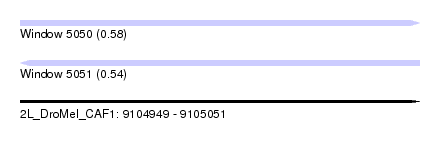
| Location | 9,104,949 – 9,105,051 |
| Length | 102 |
| Max. P | 0.579783 |

| Location | 9,104,949 – 9,105,051 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.39 |
| Energy contribution | -17.08 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
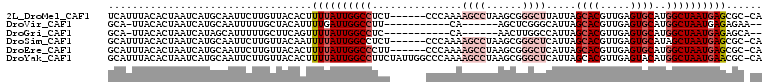
>2L_DroMel_CAF1 9104949 102 + 22407834 UCAUUUACACUAAUCAUGCAAUUCUUGUUACACUUUUAUUGGCCUCU------CCCAAAAGCCUAAGCGGGCUUAUUAGCACGUUGAGUGCAUGGCUAAUGAGCGC-CA .................(((((....)))......(((((((((...------.....((((((....))))))....((((.....))))..)))))))))))..-.. ( -24.80) >DroVir_CAF1 37457 89 + 1 GCA-UUACACUAAUCAUGCAAUUUUUGCUACAUUUUGAUUGGCCUU-----------CA------AGCUCGGGCAUUAGCACGUUGAGUGCAUGGCUAAUGAGAGAA-- (((-(..........)))).............((((.(((((((..-----------..------.((....))....((((.....))))..))))))).))))..-- ( -21.50) >DroGri_CAF1 38617 89 + 1 GCA-UUACACUAAUCAUAGCAUUUUUGCUUCAGUUUUAUUGGCCUC-----------CA------AACUUGGCCAUUAGCACGUUGAGUGCAUGGCUAAUGAGAGCA-- ((.-.............((((....))))....(((((((((((.(-----------((------....)))......((((.....))))..))))))))))))).-- ( -25.20) >DroSim_CAF1 7218 102 + 1 GCAUUUACACUAAUCAUGCAAUUCUUGUUACAAUUUUAUUGGCCUCU------CCCAAAAGCCUAAGCGGGCUCAUUAGCACGUUGAGUGCAUAGCUAAUGAGCGC-CA ((((...........))))...................((((.....------.))))..((....))((((((((((((..((.....))...)))))))))).)-). ( -23.70) >DroEre_CAF1 7317 102 + 1 GCAUUUACACUAAUCAUGCAAUUCUUGUUACACUUUUAUUGGCCCUU------CCCAAAAGCCUAAGCGGGCUCAUUAGCACGUUGAGUGCAUGGCUAAUGAGCGC-CA ((((...........))))...................((((.....------.))))..((....))((((((((((((.(((.......))))))))))))).)-). ( -24.80) >DroYak_CAF1 7630 108 + 1 GCAUUUACACUAAUCAUGCAAUUCUUGUUACACUUUUAUUGGCCUUCUAUUGGCCCAAAAGCCUAAGCGGGCUCAUUAGCACGUUGAGUACAUGGCUAAUGAACGC-CA ((((...........))))......((((...(((((...((((.......)))).)))))....))))(((((((((((.(((.......)))))))))))..))-). ( -25.80) >consensus GCAUUUACACUAAUCAUGCAAUUCUUGUUACACUUUUAUUGGCCUCU______CCCAAAAGCCUAAGCGGGCUCAUUAGCACGUUGAGUGCAUGGCUAAUGAGCGC_CA ..................................((((((((((...............((((......)))).....((((.....))))..))))))))))...... (-15.39 = -17.08 + 1.70)
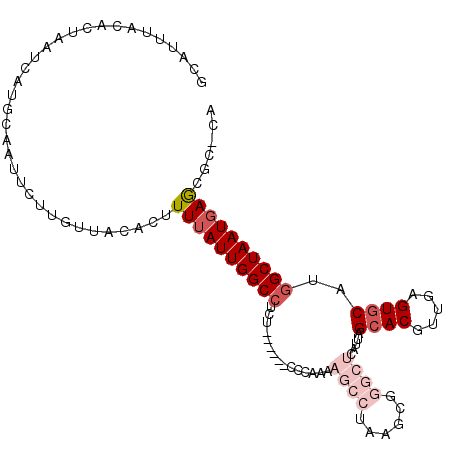

| Location | 9,104,949 – 9,105,051 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.88 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
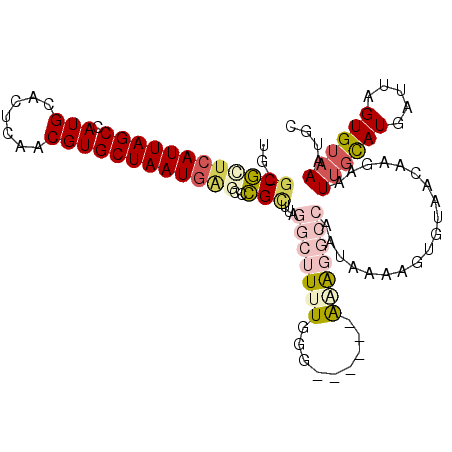
>2L_DroMel_CAF1 9104949 102 - 22407834 UG-GCGCUCAUUAGCCAUGCACUCAACGUGCUAAUAAGCCCGCUUAGGCUUUUGGG------AGAGGCCAAUAAAAGUGUAACAAGAAUUGCAUGAUUAGUGUAAAUGA ((-((.((((..((((..((((.....))))...((((....))))))))..))))------....)))).(((..((((((......))))))..))).......... ( -28.00) >DroVir_CAF1 37457 89 - 1 --UUCUCUCAUUAGCCAUGCACUCAACGUGCUAAUGCCCGAGCU------UG-----------AAGGCCAAUCAAAAUGUAGCAAAAAUUGCAUGAUUAGUGUAA-UGC --......(((((..((((((.......(((((......(((((------..-----------..)))...))......))))).....))))))..)))))...-... ( -18.30) >DroGri_CAF1 38617 89 - 1 --UGCUCUCAUUAGCCAUGCACUCAACGUGCUAAUGGCCAAGUU------UG-----------GAGGCCAAUAAAACUGAAGCAAAAAUGCUAUGAUUAGUGUAA-UGC --.((...(((((..(((((((.....))))...(((((.....------..-----------..)))))..........((((....)))))))..)))))...-.)) ( -21.00) >DroSim_CAF1 7218 102 - 1 UG-GCGCUCAUUAGCUAUGCACUCAACGUGCUAAUGAGCCCGCUUAGGCUUUUGGG------AGAGGCCAAUAAAAUUGUAACAAGAAUUGCAUGAUUAGUGUAAAUGC .(-(.((((((((((.(((.......)))))))))))))))((...((((((....------.))))))...................((((((.....))))))..)) ( -31.20) >DroEre_CAF1 7317 102 - 1 UG-GCGCUCAUUAGCCAUGCACUCAACGUGCUAAUGAGCCCGCUUAGGCUUUUGGG------AAGGGCCAAUAAAAGUGUAACAAGAAUUGCAUGAUUAGUGUAAAUGC .(-(.((((((((((.(((.......)))))))))))))))(.(((.(((((((((------.....))..))))))).)))).....((((((.....)))))).... ( -30.30) >DroYak_CAF1 7630 108 - 1 UG-GCGUUCAUUAGCCAUGUACUCAACGUGCUAAUGAGCCCGCUUAGGCUUUUGGGCCAAUAGAAGGCCAAUAAAAGUGUAACAAGAAUUGCAUGAUUAGUGUAAAUGC .(-(.((((((((((.((((.....))))))))))))))))(.(((.(((((((((((.......))))..))))))).)))).....((((((.....)))))).... ( -36.40) >consensus UG_GCGCUCAUUAGCCAUGCACUCAACGUGCUAAUGAGCCCGCUUAGGCUUUUGGG______AAAGGCCAAUAAAAGUGUAACAAGAAUUGCAUGAUUAGUGUAAAUGC ...((((((((((((.(((.......))))))))))))..)))...(((((((.........)))))))...................((((((.....)))))).... (-15.18 = -16.88 + 1.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:14 2006