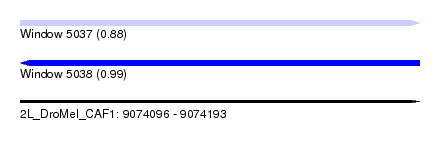
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,074,096 – 9,074,193 |
| Length | 97 |
| Max. P | 0.987257 |

| Location | 9,074,096 – 9,074,193 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
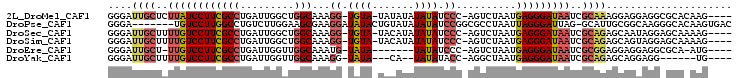
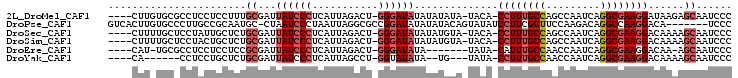
>2L_DroMel_CAF1 9074096 97 + 22407834 GGGAUUGCUCUUAUCCUUCGCCUGAUUGGCUGGCAAAGG-UGUA-UAUAUAUAUAUCCC-AGUCUAAUGAGGGAUAAUCGCAAAGGAGGAGGCGCACAAG---- .....(((.((..((((((.(((.(((((((((....((-((((-(....)))))))))-)).))))).)))(.....).....)))))))).)))....---- ( -28.60) >DroPse_CAF1 86104 96 + 1 GGGA-------UGUCCUUGGCCUGUCUUGGAAGCGAAGGAUAUACUGUAUAUAUAUCCGGCGCCUAAUUAGGGAUUAG-GCAUUGCGGCAAGGGCACAAGUGAC ....-------.((((((((((...((((...((((.(((((((.......)))))))...((((((((...))))))-)).))))..))))))).)))).))) ( -34.00) >DroSec_CAF1 67817 97 + 1 GGGAUUGCUUUUGUCCUUCGCCUGAUUGGCUGGCAAAGG-UGUA-UACAUAUAUAUCCC-AGUCUAAUGAGGGAUAAUCGCAGAGCAAUAGGAGCAAAAG---- ....((((((((((..(((.(((.(((((((((....((-((((-(....)))))))))-)).))))).)))(.....)...)))..))))))))))...---- ( -28.90) >DroSim_CAF1 73242 97 + 1 GGGAUUGCUUUUGUCCUUCGCCUGAUUGGCUGGCAAAGG-UGUA-UACAUAUAUAUCCC-AGUCUAAUGAGGGAUAAUCGCAGAGCAGUAGGAGCAAAAG---- ...((((((((((....((.(((.(((((((((....((-((((-(....)))))))))-)).))))).)))))....)).))))))))...........---- ( -28.30) >DroEre_CAF1 70158 89 + 1 GGGAUUGCU-UUGUCCUUCGCCUGAUUGGUUGGCAAAUG-UAUA-------UAUAUCCC-AGUCUAAUGAGGGAUAAUCGCGGAGGAGGAGGCGCA-AUG---- ......(((-((.((((((((((.(((((((((...(((-(...-------.)))).))-)).))))).)).(.....))))))))).)))))...-...---- ( -29.50) >DroYak_CAF1 70172 87 + 1 GGGAUUGCUUUUGUCCUUCGCCUGAUUGGUUGGCAAAGG-UAUA---CA--UAUAUACC-AGGCUAAUGAGGGAUAAUCGCAGAGCAGGAGG------UG---- ....((((..((((((((((((.....)))((((...((-((((---..--..))))))-..))))..)))))))))..)))).........------..---- ( -28.50) >consensus GGGAUUGCUUUUGUCCUUCGCCUGAUUGGCUGGCAAAGG_UAUA_UACAUAUAUAUCCC_AGUCUAAUGAGGGAUAAUCGCAGAGCAGGAGGAGCA_AAG____ ....((((..((((((((((((.........)))...((.((((.......)))).))..........)))))))))..))))..................... (-16.40 = -17.02 + 0.62)
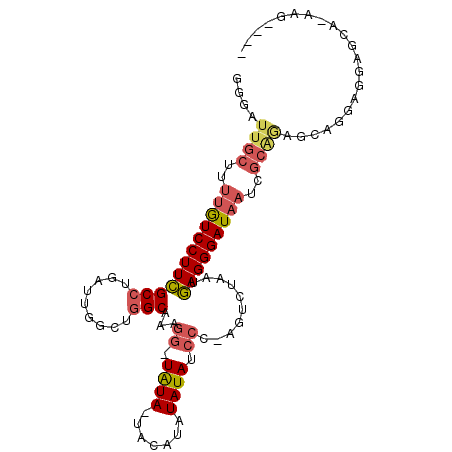
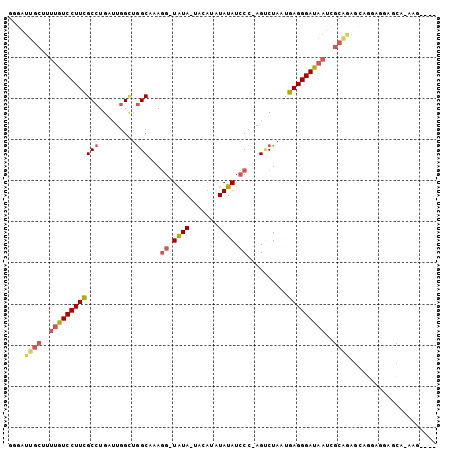
| Location | 9,074,096 – 9,074,193 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -14.31 |
| Energy contribution | -15.33 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
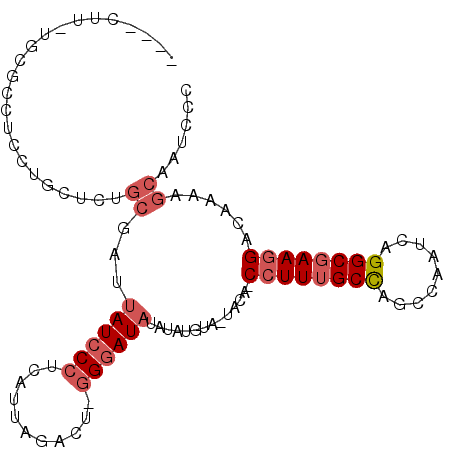
>2L_DroMel_CAF1 9074096 97 - 22407834 ----CUUGUGCGCCUCCUCCUUUGCGAUUAUCCCUCAUUAGACU-GGGAUAUAUAUAUA-UACA-CCUUUGCCAGCCAAUCAGGCGAAGGAUAAGAGCAAUCCC ----....(((..((..((((((((((.((((((..........-))))))........-....-...))))..(((.....)))))))))..)).)))..... ( -23.69) >DroPse_CAF1 86104 96 - 1 GUCACUUGUGCCCUUGCCGCAAUGC-CUAAUCCCUAAUUAGGCGCCGGAUAUAUAUACAGUAUAUCCUUCGCUUCCAAGACAGGCCAAGGACA-------UCCC (((.((((.(((((((..((...((-(((((.....)))))))...((((((((.....))))))))...))...))))...)))))))))).-------.... ( -34.20) >DroSec_CAF1 67817 97 - 1 ----CUUUUGCUCCUAUUGCUCUGCGAUUAUCCCUCAUUAGACU-GGGAUAUAUAUGUA-UACA-CCUUUGCCAGCCAAUCAGGCGAAGGACAAAAGCAAUCCC ----...........((((((.((((..((((((..........-))))))....))))-....-((((((((.........)))))))).....))))))... ( -25.40) >DroSim_CAF1 73242 97 - 1 ----CUUUUGCUCCUACUGCUCUGCGAUUAUCCCUCAUUAGACU-GGGAUAUAUAUGUA-UACA-CCUUUGCCAGCCAAUCAGGCGAAGGACAAAAGCAAUCCC ----...(((((..........((((..((((((..........-))))))....))))-....-((((((((.........)))))))).....))))).... ( -24.80) >DroEre_CAF1 70158 89 - 1 ----CAU-UGCGCCUCCUCCUCCGCGAUUAUCCCUCAUUAGACU-GGGAUAUA-------UAUA-CAUUUGCCAACCAAUCAGGCGAAGGACAA-AGCAAUCCC ----.((-(((...(((...........((((((..........-))))))..-------....-..((((((.........)))))))))...-.)))))... ( -20.80) >DroYak_CAF1 70172 87 - 1 ----CA------CCUCCUGCUCUGCGAUUAUCCCUCAUUAGCCU-GGUAUAUA--UG---UAUA-CCUUUGCCAACCAAUCAGGCGAAGGACAAAAGCAAUCCC ----..------.....((((.((((..(((.((..........-)).)))..--))---))..-((((((((.........)))))))).....))))..... ( -18.20) >consensus ____CUU_UGCGCCUCCUGCUCUGCGAUUAUCCCUCAUUAGACU_GGGAUAUAUAUGUA_UACA_CCUUUGCCAGCCAAUCAGGCGAAGGACAAAAGCAAUCCC .......................((...((((((...........))))))..............((((((((.........))))))))......))...... (-14.31 = -15.33 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:01 2006