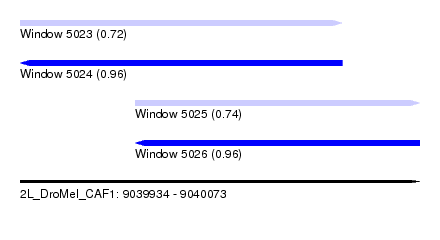
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,039,934 – 9,040,073 |
| Length | 139 |
| Max. P | 0.961226 |

| Location | 9,039,934 – 9,040,046 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
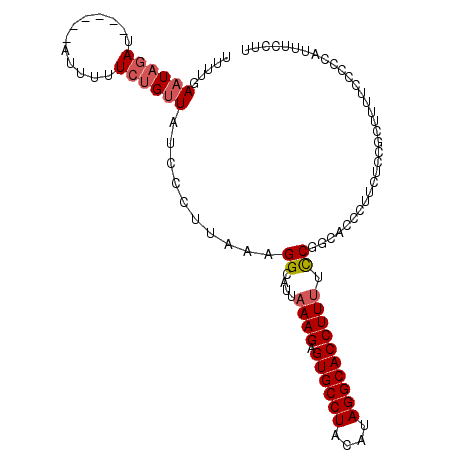
>2L_DroMel_CAF1 9039934 112 + 22407834 ACUAAUGUGGGCAUAAUAGCUUUCGAUUAUCGAGAAGAAGAAGGAAAUGGGGGAAAAGCGGAGAAGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAG .....((.((((((......(((((.....)))))...........................((((((((((.....((....)).....)))))))))).)))))).)).. ( -28.10) >DroSec_CAF1 34009 112 + 1 ACUAAUGUGGGCAUAAUAGCUUUCGAUUAUCGAGGAGAAGAAGGAAAUGGGGGAAAAGCGGAGAGGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAG .....((.((((((....((((((.(((.((...........)).))).))....))))...((((((((((.....((....)).....)))))))))).)))))).)).. ( -27.80) >DroSim_CAF1 37790 111 + 1 ACUAAUGUGGGCAUAAUAGCUUUCGAUUAUCGAGGAGAAGAAGGAAAUGGGGGAAAAGCGGAGAA-GGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAG .....((.((((((......(((((.....))))).(((((.................(((....-....)))......((((((....))))))))))).)))))).)).. ( -25.70) >DroEre_CAF1 36468 111 + 1 ACUAAUGUGGGCGUAAUAGCUUUCGAUUAUCGGGGGGAAGAAGGAAAAGAGGGCGAGGAGGAGGAGCCUG-CGAAGAAGGUGCCUGUAUAGGCACUCUUUAAUGCCUUUAAG ...................((..((.....))..))............(((((((..((((((..((((.-......))))((((....)))).))))))..)))))))... ( -31.60) >DroYak_CAF1 35260 112 + 1 ACUAAUGUGGGCAUAAUAGCUUUCGAUUAUCGAGGGGCAGAAAGAAAAGAGGGUAAGGCGGAGGAGCCUGGUGGAAAAGGUGCCUCUAUAGGCACUCUUUAAUGCCUUUAAG .....((.((((((.....((((((.....)))))).........((((((.((...(..((((.((((........)))).))))..)..)).)))))).)))))).)).. ( -34.60) >consensus ACUAAUGUGGGCAUAAUAGCUUUCGAUUAUCGAGGAGAAGAAGGAAAUGGGGGAAAAGCGGAGAAGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAG .....((.((((((.....(((((..((......))...)))))...............................((((((((((....)))))).)))).)))))).)).. (-23.08 = -22.20 + -0.88)


| Location | 9,039,934 – 9,040,046 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9039934 112 - 22407834 CUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCUUCUCCGCUUUUCCCCCAUUUCCUUCUUCUUCUCGAUAAUCGAAAGCUAUUAUGCCCACAUUAGU ......(((((.((((.((((((....))))))))))................((((((.............................))))))....)))))......... ( -19.85) >DroSec_CAF1 34009 112 - 1 CUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCCUCUCCGCUUUUCCCCCAUUUCCUUCUUCUCCUCGAUAAUCGAAAGCUAUUAUGCCCACAUUAGU ......(((((.((((.((((((....))))))))))................((((((.............................))))))....)))))......... ( -19.85) >DroSim_CAF1 37790 111 - 1 CUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACC-UUCUCCGCUUUUCCCCCAUUUCCUUCUUCUCCUCGAUAAUCGAAAGCUAUUAUGCCCACAUUAGU ......(((((.((((.((((((....))))))))))..(((....-....))).........................(((.....)))........)))))......... ( -19.90) >DroEre_CAF1 36468 111 - 1 CUUAAAGGCAUUAAAGAGUGCCUAUACAGGCACCUUCUUCG-CAGGCUCCUCCUCCUCGCCCUCUUUUCCUUCUUCCCCCCGAUAAUCGAAAGCUAUUACGCCCACAUUAGU ......(((....(((.((((((....)))))))))....(-(.(((...........)))...................((.....))...))......)))......... ( -20.30) >DroYak_CAF1 35260 112 - 1 CUUAAAGGCAUUAAAGAGUGCCUAUAGAGGCACCUUUUCCACCAGGCUCCUCCGCCUUACCCUCUUUUCUUUCUGCCCCUCGAUAAUCGAAAGCUAUUAUGCCCACAUUAGU ......((((..(((((((((((....))))))..........((((......))))..........))))).))))..(((.....)))..((......)).......... ( -24.80) >consensus CUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCUUCUCCGCUUUUCCCCCAUUUCCUUCUUCUCCUCGAUAAUCGAAAGCUAUUAUGCCCACAUUAGU ......(((((.((((.((((((....))))))))))..........................................(((.....)))........)))))......... (-16.84 = -17.44 + 0.60)


| Location | 9,039,974 – 9,040,073 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9039974 99 + 22407834 AAGGAAAUGGGGGAAAAGCGGAGAAGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAGGGAUAACAGAAAAAU------AUCUAUUCAAAA ...(((.......(((.(((..((((((((((.....((....)).....))))))))))..))).)))...(((((.........)------)))).))).... ( -23.20) >DroSec_CAF1 34049 99 + 1 AAGGAAAUGGGGGAAAAGCGGAGAGGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAGGGAUAACAGAAAAAU------AUCUAUUCAAAA ...(((.((((((...(..(((((((((..((......))..))).((....))))))))..).))))))..(((((.........)------)))).))).... ( -23.30) >DroSim_CAF1 37830 98 + 1 AAGGAAAUGGGGGAAAAGCGGAGAA-GGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAGGGAUAACAGAAAAAU------AUCUAUUCAAAA ...(((.((((((.....(((....-....)))..((((((((((....)))))).))))....))))))..(((((.........)------)))).))).... ( -21.90) >DroEre_CAF1 36508 104 + 1 AAGGAAAAGAGGGCGAGGAGGAGGAGCCUG-CGAAGAAGGUGCCUGUAUAGGCACUCUUUAAUGCCUUUAAGGGGUAACAGAAGUAUACUCGUAUCUAAUCAAGC ..(((...(((((((..((((((..((((.-......))))((((....)))).))))))..)))))))...(((((((....)).)))))...)))........ ( -30.60) >DroYak_CAF1 35300 99 + 1 AAAGAAAAGAGGGUAAGGCGGAGGAGCCUGGUGGAAAAGGUGCCUCUAUAGGCACUCUUUAAUGCCUUUAAGGGGUAACCGAAAUAU------AUAUAUUAAAGA ........(((((((((((......))))......((((((((((....)))))).))))..)))))))..((.....)).......------............ ( -26.80) >consensus AAGGAAAUGGGGGAAAAGCGGAGAAGGGUGCCGGAAAAGGUGCCUAUGUAGGCACUCUUUAAUGCCUUUAAGGGAUAACAGAAAAAU______AUCUAUUCAAAA ........(((((.....(((.........)))..((((((((((....)))))).))))....))))).................................... (-14.62 = -14.66 + 0.04)


| Location | 9,039,974 – 9,040,073 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -20.44 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9039974 99 - 22407834 UUUUGAAUAGAU------AUUUUUCUGUUAUCCCUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCUUCUCCGCUUUUCCCCCAUUUCCUU .....((((((.------.....)))))).......((((((...((((.((((((....))))))))))...((.........))))))))............. ( -19.80) >DroSec_CAF1 34049 99 - 1 UUUUGAAUAGAU------AUUUUUCUGUUAUCCCUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCCUCUCCGCUUUUCCCCCAUUUCCUU .....((((((.------.....)))))).......((((((...((((.((((((....))))))))))...((....)).....))))))............. ( -20.30) >DroSim_CAF1 37830 98 - 1 UUUUGAAUAGAU------AUUUUUCUGUUAUCCCUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACC-UUCUCCGCUUUUCCCCCAUUUCCUU .....((((((.------.....)))))).......((((((...((((.((((((....))))))))))...((....-....))))))))............. ( -20.60) >DroEre_CAF1 36508 104 - 1 GCUUGAUUAGAUACGAGUAUACUUCUGUUACCCCUUAAAGGCAUUAAAGAGUGCCUAUACAGGCACCUUCUUCG-CAGGCUCCUCCUCCUCGCCCUCUUUUCCUU (((((....((((.(((....))).)))).........(((((((....)))))))...))))).........(-((((........))).))............ ( -20.60) >DroYak_CAF1 35300 99 - 1 UCUUUAAUAUAU------AUAUUUCGGUUACCCCUUAAAGGCAUUAAAGAGUGCCUAUAGAGGCACCUUUUCCACCAGGCUCCUCCGCCUUACCCUCUUUUCUUU ............------.......(((...........((....((((.((((((....))))))))))....))((((......)))).)))........... ( -20.90) >consensus UUUUGAAUAGAU______AUUUUUCUGUUAUCCCUUAAAGGCAUUAAAGAGUGCCUACAUAGGCACCUUUUCCGGCACCCUUCUCCGCUUUUCCCCCAUUUCCUU .....((((((............))))))..........((....((((.((((((....)))))))))).))................................ (-13.12 = -13.76 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:50 2006