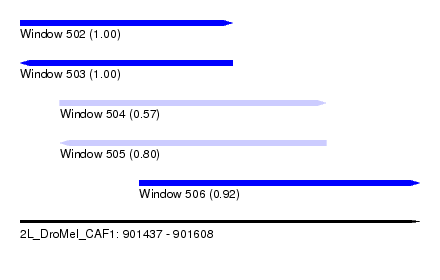
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 901,437 – 901,608 |
| Length | 171 |
| Max. P | 0.999958 |

| Location | 901,437 – 901,528 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 58.24 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 901437 91 + 22407834 AAGGCAGGCUACAUAUAUGAAAAUAAUGAUACUUUUAAAAUUUAUUGCAGAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAU ......((...(((.(((....))))))........................(((((((.((((....))))..)))))))..))...... ( -21.30) >DroSim_CAF1 27788 91 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAUUUAUUGCAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAU ....................................................(((((((.((((....))))..))))))).......... ( -18.20) >DroYak_CAF1 30929 91 + 1 CAAUAAUUAAUAUAAUUGAGACCUUAGAGCUACUUUUAAAUUUAUUUUUUAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAU ..((((..((((...((((((............))))))...))))..))))(((((((.((((....))))..))))))).......... ( -20.40) >consensus _A___A________AU____A__U_A_______UUU_AAAUUUAUUGCAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAU ....................................................(((((((.((((....))))..))))))).......... (-18.20 = -18.20 + -0.00)

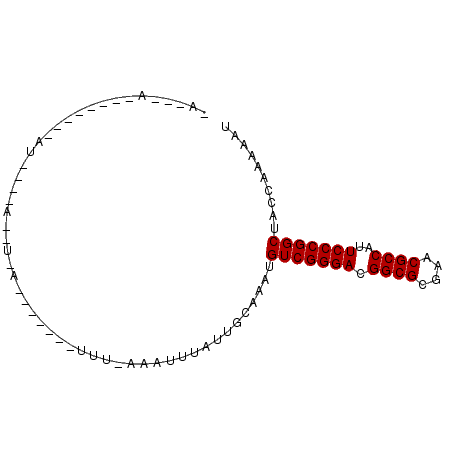
| Location | 901,437 – 901,528 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 58.24 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 901437 91 - 22407834 AUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUCUGCAAUAAAUUUUAAAAGUAUCAUUAUUUUCAUAUAUGUAGCCUGCCUU ......(((((.(((((.(((((....))))))))))....(((((..........((((((....))))))......))))).))))).. ( -26.09) >DroSim_CAF1 27788 91 - 1 AUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUUUGCAAUAAAUNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .......((((.(((((.(((((....))))))))))....)))).............................................. ( -17.90) >DroYak_CAF1 30929 91 - 1 AUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUAAAAAAUAAAUUUAAAAGUAGCUCUAAGGUCUCAAUUAUAUUAAUUAUUG ..((((((.((((((((.(((((....))))))))))...((((.......))))......)))))))))..................... ( -21.50) >consensus AUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUAUGCAAUAAAUUU_AAA_______U_A__U____AU________U___U_ ..........(.(((((.(((((....)))))))))).).................................................... (-17.60 = -17.60 + -0.00)

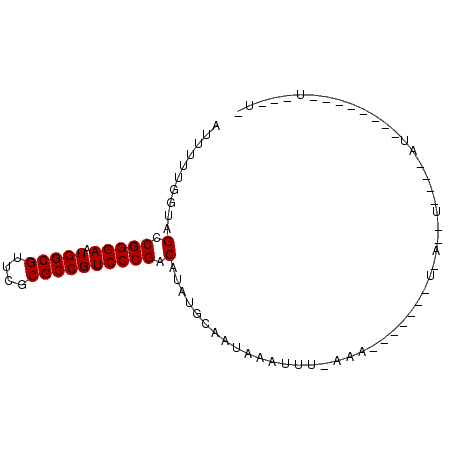
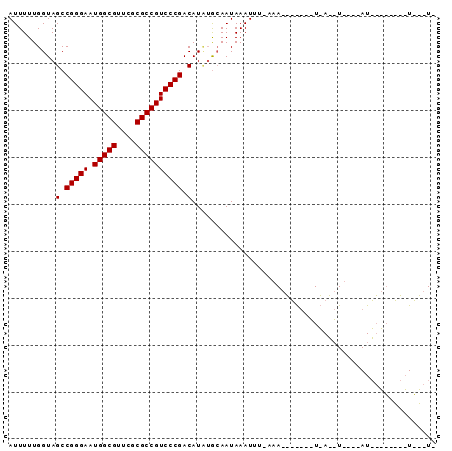
| Location | 901,454 – 901,568 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 901454 114 + 22407834 UGAAAAUAAUGAUACUUUUAAAAUUUAUUGCAGAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGG ...........................((((....(((((((.((((....))))..)))))))...................((.((((((.......))))))))))))... ( -26.10) >DroSim_CAF1 27805 114 + 1 NNNNNNNNNNNNNNNNNNNNNNAUUUAUUGCAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGG ...........................((((....(((((((.((((....))))..)))))))...................((.((((((.......))))))))))))... ( -26.10) >DroEre_CAF1 37022 104 + 1 ----------AACUAAUUUGAAAUGUAUUUUCCAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGG ----------......((((...((((((((....(((((((.((((....))))..))))))).....)))).))))....(((.((((((.......))))))))))))).. ( -26.40) >DroYak_CAF1 30946 114 + 1 GAGACCUUAGAGCUACUUUUAAAUUUAUUUUUUAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGG ....((((...((......((((.......)))).(((((((.((((....))))..)))))))...............)).(((.((((((.......)))))))))..)))) ( -27.20) >consensus __________AACUA_UUU_AAAUUUAUUGCAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGG ...................................(((((((.((((....))))..)))))))..................(((.((((((.......)))))))))...... (-23.30 = -23.30 + 0.00)

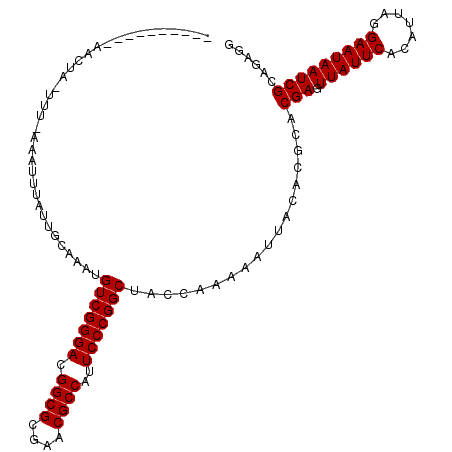
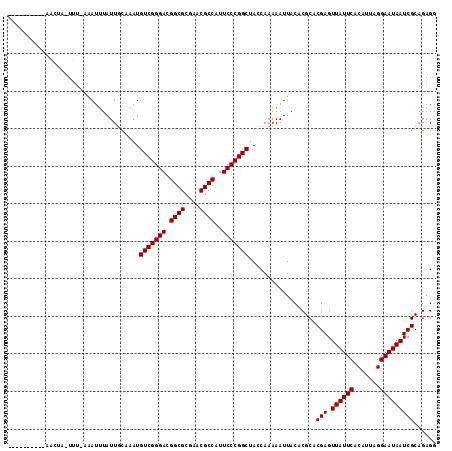
| Location | 901,454 – 901,568 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -26.23 |
| Energy contribution | -27.35 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 901454 114 - 22407834 CCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUCUGCAAUAAAUUUUAAAAGUAUCAUUAUUUUCA .....(((((((((((....).)))))).))))((.((.(((((...((((.(((((.(((((....))))))))))....))))...))))).))...))............. ( -29.80) >DroSim_CAF1 27805 114 - 1 CCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUUUGCAAUAAAUNNNNNNNNNNNNNNNNNNNNNN .....(((((((((((....).)))))).))))...(((((....((.....(((((.(((((....)))))))))).)).)))))............................ ( -26.50) >DroEre_CAF1 37022 104 - 1 CCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUGGAAAAUACAUUUCAAAUUAGUU---------- .....(((((((((((....).)))))).))))..(((((.(((((.((.(.(((((.(((((....)))))))))).))).))))))))))............---------- ( -31.50) >DroYak_CAF1 30946 114 - 1 CCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUAAAAAAUAAAUUUAAAAGUAGCUCUAAGGUCUC .....(((((((((((....).)))))).)))).........((((((.((((((((.(((((....))))))))))...((((.......))))......))))))))).... ( -29.00) >consensus CCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGCCGUCCCGACAUAUAAAAUAAAUUU_AAA_UAGUU__________ .....(((((((((((....).)))))).)))).......(((((((((.(.(((((.(((((....)))))))))).)))))))))).......................... (-26.23 = -27.35 + 1.12)


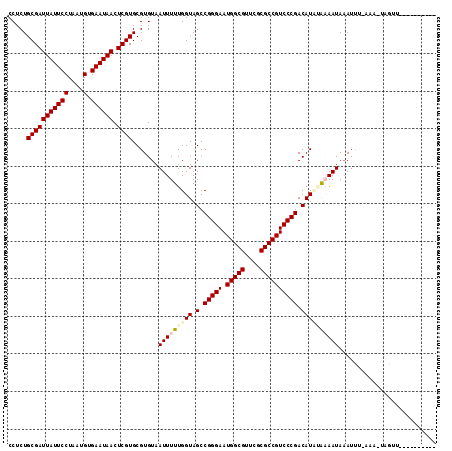
| Location | 901,488 – 901,608 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -38.04 |
| Energy contribution | -38.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 901488 120 + 22407834 UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAGGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((.(((.((((((.......)))))))))..(((.....))).)).(((((((....))...))))).....)). ( -38.40) >DroSec_CAF1 30189 120 + 1 UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAGGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((.(((.((((((.......)))))))))..(((.....))).)).(((((((....))...))))).....)). ( -38.40) >DroSim_CAF1 27839 120 + 1 UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAAGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((((((.((((((.......))))))))((.(((.....))).))....))))...(((...))).......)). ( -38.20) >DroEre_CAF1 37046 120 + 1 UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAGGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((.(((.((((((.......)))))))))..(((.....))).)).(((((((....))...))))).....)). ( -38.40) >DroYak_CAF1 30980 120 + 1 UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAGGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((.(((.((((((.......)))))))))..(((.....))).)).(((((((....))...))))).....)). ( -38.40) >consensus UGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAGGUGCAAUGGCCAAGCCUCACUCCGGA .(((((((.((((....))))..)))))))..((...........((((((.((((((.......))))))))((.(((.....))).))....))))...(((...))).......)). (-38.04 = -38.04 + -0.00)

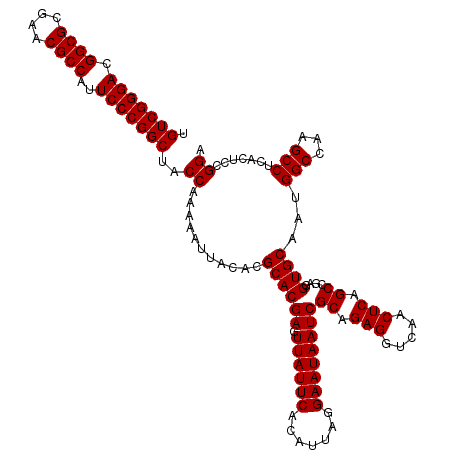
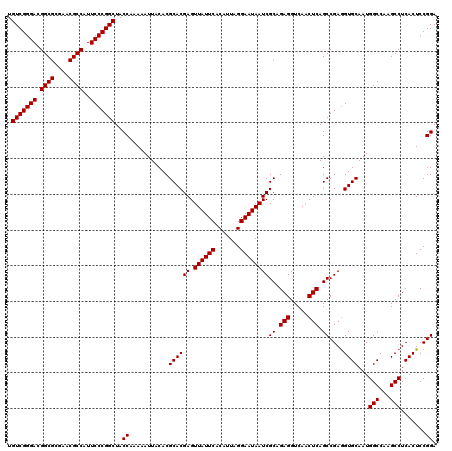
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:52 2006