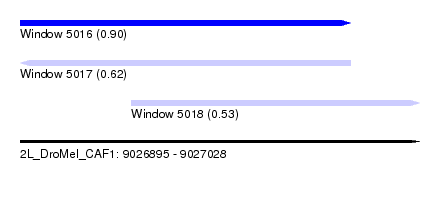
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,026,895 – 9,027,028 |
| Length | 133 |
| Max. P | 0.903634 |

| Location | 9,026,895 – 9,027,005 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9026895 110 + 22407834 AGGUUUUCCACCC--ACUGUCAGCAGCACAUC-AGAUGUCUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCU .((....))....--(((((..((((....((-((((......(((((((.(((((....)))))...))))))).....))))))....))))..)))))............ ( -24.50) >DroVir_CAF1 45264 100 + 1 ---CACUGCCAUUUGGCUG---GCAGCCCUCGU-------UUCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCU ---.((((((....)))((---((((.......-------...(((((((.(((((....)))))...))))))).(((....)))....))))))..)))............ ( -21.50) >DroPse_CAF1 24528 105 + 1 AUAUACUCCAUCCUGGCUGUCAGCAG-ACAUC-------UUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCU .............(((((.......(-(((..-------.....)))).......)))))...((((((.(((...(((....)))..(((((....)))))))).)))))). ( -23.84) >DroWil_CAF1 44048 106 + 1 -ACUUUU---CCU-ACUUGUCAGCUACAUGGC--GACAGCUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCU -......---...-...((((.(((....)))--))))((...(((((((.(((((....)))))...))))))).........(((((.(((....))))))))..)).... ( -21.90) >DroMoj_CAF1 39397 110 + 1 ---AAAAGUCAUUUGGGUGAAAGCAGCUCCCGUUGCCUGCUUCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCU ---....((((..(...((((((((((.......).)))))).(((((((.(((((....)))))...))))))).))).)..)))).(((((....)))))........... ( -24.20) >DroPer_CAF1 24475 105 + 1 AUAUACUCCAUCCUGGCUGUCAGCAG-ACAUC-------UUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAGCAGUUGUUUUGCAUCU .............(((((.......(-(((..-------.....)))).......)))))...((((((.(((...(((....)))..(((((....)))))))).)))))). ( -23.84) >consensus ___UACUCCAACCUGGCUGUCAGCAGCACAUC_______CUCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACAGUUGUUUUGCAUCU ......................((((.................(((((((.(((((....)))))...))))))).........(((((.(((....)))))))))))).... (-16.07 = -16.10 + 0.03)

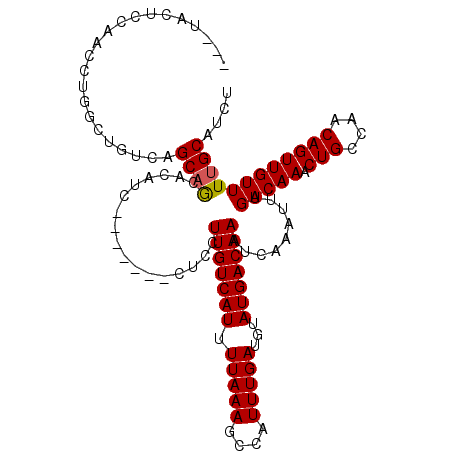
| Location | 9,026,895 – 9,027,005 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9026895 110 - 22407834 AGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAGACAUCU-GAUGUGCUGCUGACAGU--GGGUGGAAAACCU ............(((((..(((((((((((..(((((...(((((.......))))).))))).))))))(....)....-.....)))))..)))))--((((.....)))) ( -32.90) >DroVir_CAF1 45264 100 - 1 AGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGAA-------ACGAGGGCUGC---CAGCCAAAUGGCAGUG--- ...(((..(((....))).))).(((((((..(((((...(((((.......))))).))))).)))))))..-------......(((((---((......))))))).--- ( -25.50) >DroPse_CAF1 24528 105 - 1 AGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAA-------GAUGU-CUGCUGACAGCCAGGAUGGAGUAUAU ..((((....((..(.(((((..(((((((..(((((...(((((.......))))).))))).)))))))...-------..(((-(....))))))))).)))..)))).. ( -25.30) >DroWil_CAF1 44048 106 - 1 AGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAGCUGUC--GCCAUGUAGCUGACAAGU-AGG---AAAAGU- .............((((((((..(((((((..(((((...(((((.......))))).))))).)))))))..)))(((--((......)).))).)))-)).---......- ( -24.20) >DroMoj_CAF1 39397 110 - 1 AGAUGCAAAACAACUGUUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGAAGCAGGCAACGGGAGCUGCUUUCACCCAAAUGACUUUU--- .((.(((...(..((((((((..(((((((..(((((...(((((.......))))).))))).)))))))..))...))))))..).)))..))...............--- ( -23.10) >DroPer_CAF1 24475 105 - 1 AGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAA-------GAUGU-CUGCUGACAGCCAGGAUGGAGUAUAU ..((((....((..(.(((((..(((((((..(((((...(((((.......))))).))))).)))))))...-------..(((-(....))))))))).)))..)))).. ( -25.30) >consensus AGAUGCAAAACAACUGCUGGCAGUUUGUCAAAUUUGAUUUGUCAUACAUCAAAUGGCUUUAAAAUGACAAGGAG_______GAUGUGCUGCUGACAGCCAAGAUGGAGUA___ .............(((..((((((((((((..(((((...(((((.......))))).))))).)))))))................)))))..)))................ (-15.30 = -15.82 + 0.53)

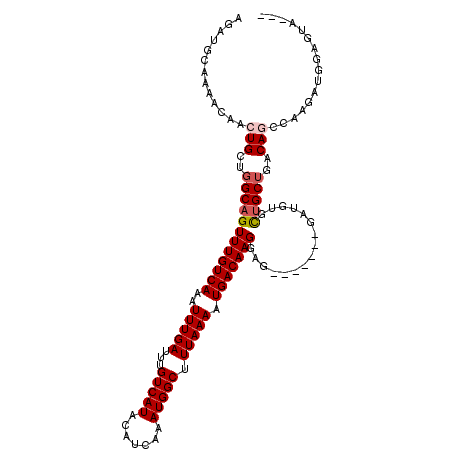
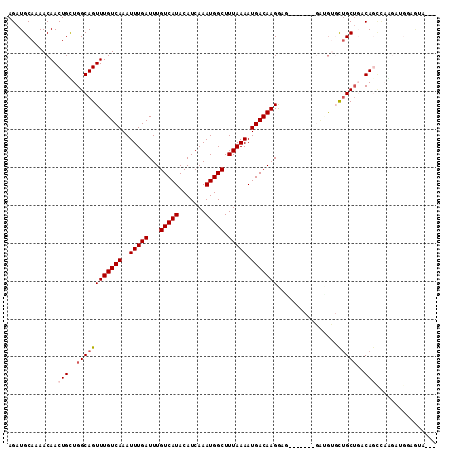
| Location | 9,026,932 – 9,027,028 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9026932 96 + 22407834 UCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG-------CCAACAGUUGUUUUGCAUCUUUGCAACAUUUUUCACAGCA------GCC----------- ...(((((((.(((((....)))))...)))))))...............(((-------(........((.(((((....))))).))........)))------)..----------- ( -17.29) >DroVir_CAF1 45291 96 + 1 UUCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG-------CCAACAGUUGUUUUGCAUCUUUGCAACAUUUUUCAACGCC------UUC----------- ...(((((((.(((((....)))))...))))))).......((((..(((((-------....)))))...(((((....)))))......))))....------...----------- ( -16.80) >DroPse_CAF1 24560 100 + 1 UCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG-------CCAGCAGUUGUUUUGCAUCUUUGCAACAUUUUUCACAGAGGACCCUG------------- ((((((((((.(((((....)))))...)))))))......((((...(((((-------....)))))...(((((....))))).........))))))).....------------- ( -21.00) >DroGri_CAF1 32188 103 + 1 UUCAUGUCAUUUUAAAGUCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUGCCAACUGCCAACAGUUGUUUUGCAUCUUUGCAACAUUUUUCAACUCC------UCC----------- ....((((((.(((((....)))))...))))))........((((.(((.((.((((((....))))))..(((((....)))))))))).))))....------...----------- ( -18.80) >DroWil_CAF1 44081 107 + 1 UCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG-------CCAGCAGUUGUUUUGCAUCUUUGUAGCAUUUUUCAAAACA------GAGCGCAGAGUCGA ...(((((((.(((((....)))))...))))))).......(((((.(((((-------....)))))..(((((..((((((.............)))------))).)))))))))) ( -24.72) >DroPer_CAF1 24507 102 + 1 UCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG-------CCAGCAGUUGUUUUGCAUCUUUGCAACAUUUUUCACAGAGGACCCUGCC----------- ((((((((((.(((((....)))))...)))))))......((((...(((((-------....)))))...(((((....))))).........))))))).......----------- ( -21.00) >consensus UCCUUGUCAUUUUAAAGCCAUUUGAUGUAUGACAAAUCAAAUUUGACAAACUG_______CCAACAGUUGUUUUGCAUCUUUGCAACAUUUUUCAAAGCA______GCC___________ ...(((((((.(((((....)))))...))))))).........(((((.(((...........))))))))(((((....))))).................................. (-14.21 = -14.10 + -0.11)

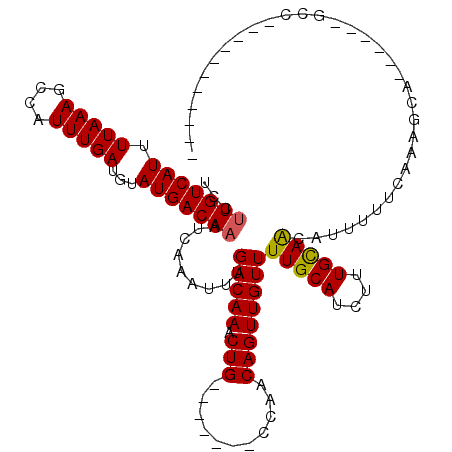
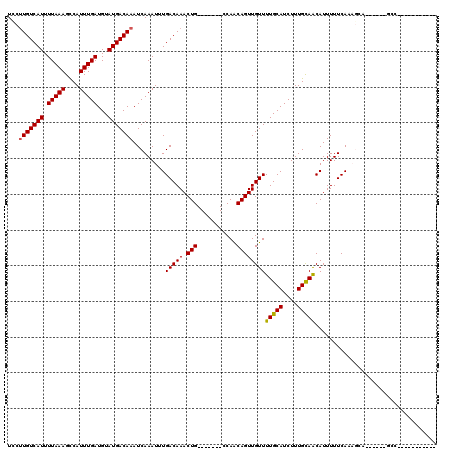
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:42 2006