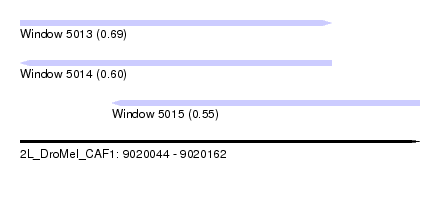
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,020,044 – 9,020,162 |
| Length | 118 |
| Max. P | 0.688831 |

| Location | 9,020,044 – 9,020,136 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -15.39 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
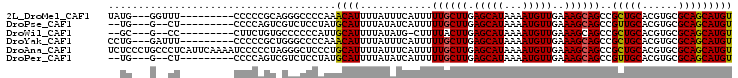
>2L_DroMel_CAF1 9020044 92 + 22407834 UAUG---GGUUU---------CCCCCGCAGGGCCCCAAACAUUUUAUUUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGCUGCACGUGCGCAGCAUGU ...(---((...---------..)))(((((((....((((((((((((((........)))).)))))))))).......))).))))((((((...)))))) ( -29.90) >DroPse_CAF1 15994 88 + 1 --UG---G--CU---------CCCCAGUCGUCUCCUAUGCAUUUUAUAUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGUUGCACGUGCGCAGCAUGU --.(---(--((---------.....((..((......(((((((((.(((........)))..))))))))).))..))))))(((((......))))).... ( -21.20) >DroWil_CAF1 28242 87 + 1 --GC---G--CC---------CUUCUGUGCCCCCCAUUGCAUUUUAUAUG-CUUUUACUUGAGCAUAAAAUGUUGAAAGCAGCCGCUGCACGUGCGCAGCAUGU --((---(--(.---------....(((((........((((((..((((-(((......)))))))))))))....(((....)))))))).))))....... ( -23.60) >DroYak_CAF1 13779 92 + 1 CCUG---GAUUU---------CCCCCGCUGGGCCCCAAACAUUUUAUUUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGCUGCACGUGCGCAGCAUGU .(((---..(((---------((((....))).....((((((((((((((........)))).)))))))))))))).)))..(((((......))))).... ( -25.30) >DroAna_CAF1 19164 104 + 1 UCUCCCUGCCCUCAUUCAAAAUCCCCCUAGGGCUCCCUGCAUUUUAUUUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGCUGCACGUGCGCAGCAUGU .......(((((................)))))...((((.(((((...(((((((((....))).)))))).)))))))))..(((((......))))).... ( -28.69) >DroPer_CAF1 15858 88 + 1 --UG---G--CU---------CCCCAGUCGUCUCCUAUGCAUUUUAUAUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGUUGCACGUGCGCAGCAUGU --.(---(--((---------.....((..((......(((((((((.(((........)))..))))))))).))..))))))(((((......))))).... ( -21.20) >consensus __UG___G__CU_________CCCCCGUAGGCCCCCAUGCAUUUUAUAUCAUUUUUGCUUGAGCAUAAAAUGUUGAAAGCAGCCGCUGCACGUGCGCAGCAUGU ......................................((((............((((((.(((((...)))))..))))))..(((((......))))))))) (-15.39 = -15.12 + -0.28)

| Location | 9,020,044 – 9,020,136 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -13.64 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9020044 92 - 22407834 ACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAAAUAAAAUGUUUGGGGCCCUGCGGGGG---------AAACC---CAUA ...(((((((....)))))))((.((((..((((((((((..(((........))).))))))))))..))))...)).(((.---------...))---)... ( -35.00) >DroPse_CAF1 15994 88 - 1 ACAUGCUGCGCACGUGCAACGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAUAUAAAAUGCAUAGGAGACGACUGGGG---------AG--C---CA-- ....(.((((....)))).)((((.(((((..(((((((((.(((........))))))))))))....(....)))..))).---------))--)---).-- ( -22.50) >DroWil_CAF1 28242 87 - 1 ACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCAAGUAAAAG-CAUAUAAAAUGCAAUGGGGGGCACAGAAG---------GG--C---GC-- ...(((((((....)))))))((((((..(..(((((((((((........))-))))..))))).....)..))).(....)---------))--)---..-- ( -25.10) >DroYak_CAF1 13779 92 - 1 ACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAAAUAAAAUGUUUGGGGCCCAGCGGGGG---------AAAUC---CAGG ...(((((((....)))))))(((((((..((((((((((..(((........))).))))))))))..))))..)))(((..---------...))---)... ( -29.80) >DroAna_CAF1 19164 104 - 1 ACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAAAUAAAAUGCAGGGAGCCCUAGGGGGAUUUUGAAUGAGGGCAGGGAGA ...(((((((....))))))).(((((((((.((((((((..(((........))).))))))))(((((..(((....))).)))))..)))))))))..... ( -34.80) >DroPer_CAF1 15858 88 - 1 ACAUGCUGCGCACGUGCAACGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAUAUAAAAUGCAUAGGAGACGACUGGGG---------AG--C---CA-- ....(.((((....)))).)((((.(((((..(((((((((.(((........))))))))))))....(....)))..))).---------))--)---).-- ( -22.50) >consensus ACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCAAGCAAAAAUGAAAUAAAAUGCAUGGGGCCCGACGGGGG_________AG__C___CA__ ...(((((((....)))))))((.........((((((.(((....))))))))).........))...................................... (-13.64 = -13.80 + 0.17)



| Location | 9,020,071 – 9,020,162 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.39 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9020071 91 - 22407834 CAG--------GCAUCCACAUU-----C-GAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCU--------CAAGCAAAAAUGAAAUAAAAUG .((--------(((.((.(...-----.-..)............((((((....)))))))).)))))....((((((.(((.--------...))))))))).......... ( -21.60) >DroVir_CAF1 23937 97 - 1 CA--------AACGUCU-UGAU-----A-CAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCUCUGAUAUUUGAGUAAAAA-CAAAUAAAAUG ..--------......(-(((.-----.-(((...........(((((((....))))))).)))...))))((((((((((((........)))).....-...)))))))) ( -19.40) >DroGri_CAF1 22528 100 - 1 CAG------AGGCAUCUAUGAU-----A-CAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUACGUCCCGAUGUUGAAGCAAAGA-CGAAUAAAAUG ...------.(((((((.....-----.-.)))).........(((((((....))))))))))((((.(((((((.........))))))))))).....-........... ( -24.00) >DroEre_CAF1 13228 99 - 1 CAGGCAUCCAGGCAUCCACAUU-----C-GAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCU--------CAAGCAAAAAUGAAAUAAAAUG .(((((.((....((((.....-----.-).)))..........((((((....)))))))).)))))....((((((.(((.--------...))))))))).......... ( -23.50) >DroWil_CAF1 28265 94 - 1 CA---------CCACCCUCAUAUCAACG-GACAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCU--------CAAGUAAAAG-CAUAUAAAAUG ..---------((..((..........)-)..............((((((....))))))))..........(((((((((((--------........))-))))..))))) ( -17.50) >DroAna_CAF1 19203 90 - 1 C----------GCAUCCACAUU-----UGGAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCU--------CAAGCAAAAAUGAAAUAAAAUG .----------(((((((....-----))))............(((((((....)))))))..)))......((((((.(((.--------...))))))))).......... ( -22.10) >consensus CA_________GCAUCCACAUU_____A_GAGAUAAUUAAACAUGCUGCGCACGUGCAGCGGCUGCUUUCAACAUUUUAUGCU________CAAGCAAAAA_CAAAUAAAAUG ...........................................(((((((....)))))))...........((((((((.........................)))))))) (-12.23 = -12.39 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:39 2006