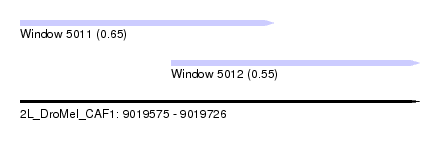
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,019,575 – 9,019,726 |
| Length | 151 |
| Max. P | 0.647532 |

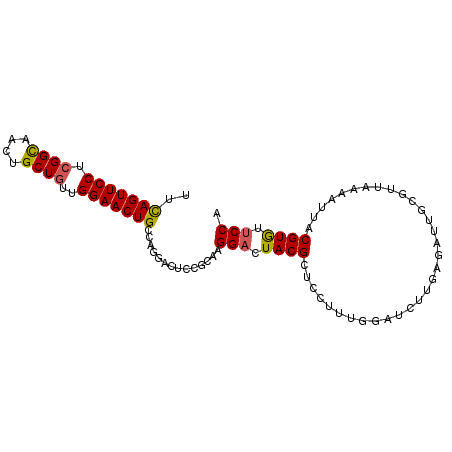
| Location | 9,019,575 – 9,019,671 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -14.56 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9019575 96 + 22407834 UCACAUGCAACAUGUUGCAG----UGUUCCAGCCGAACUGUUUUAGC-------CAGCUUAACAUUCUUUCAGUUCCUCGGCAAAUGCUGUUGGAACUGCCGGGACU- ..((.(((((....))))))----)(((((....(((.((((..((.-------...)).)))))))...(((((((.((((....))))..)))))))..))))).- ( -31.20) >DroVir_CAF1 23320 103 + 1 GUAUGUGCAACUUGUUGUAGCUCAUGUU-----AAAAAUGUCAAAGCAACAUCUCAGCUCGACUCUGGCGCAGUUUCAGAGCAGCUGCUGUGGAAACUACAACAAUUU ...........(((((((((.((.....-----.....(((....))).(((..(((((.(.((((((.......))))))))))))..)))))..)))))))))... ( -26.00) >DroSec_CAF1 13549 96 + 1 UCACAUGCAACUUGUUGCAG----UGUUCCAGCCGAACUGUUUUAGC-------CAGCUUAACACUCUUUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCGGGACG- ..((.(((((....))))))----)(((((.((.(((.((((..((.-------...)).))))....)))((((((.((((....))))..)))))))).))))).- ( -29.90) >DroSim_CAF1 14582 96 + 1 UCACAUGCAACUUGUUGCAG----UGUUCCAGCCGAACUGUUUUAGC-------CAGCUUAACACUCUUUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGACCG- .....(((((....)))))(----((((..(((.(............-------).))).)))))(((..(((((((.((((....))))..)))))))..)))...- ( -27.60) >DroEre_CAF1 12753 96 + 1 UCACAUGCAACUUGCCUUGG----UGUUCCAGCCGAACUGUUUCAGC-------CAGCUUAACACGCUUUUAGUUCCGCGGCAACUGCUGUUGGAACUGCCACGAUU- ......((.....))(.(((----(((((((((.((((((....(((-------...........)))..)))))).(((....).)).)))))))).)))).)...- ( -25.00) >DroYak_CAF1 13318 96 + 1 UCACUUACAACUUGCUUCAA----UGUUCGAGCCGAACUGCUUUAGC-------CAUCUUAACACUCGUUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGGACU- ..........((((......----.(((.((((......)))).)))-------................(((((((.((((....))))..))))))).))))...- ( -22.30) >consensus UCACAUGCAACUUGUUGCAG____UGUUCCAGCCGAACUGUUUUAGC_______CAGCUUAACACUCUUUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGGACU_ .....(((((....)))))......((((.....)))).(((..(((.........))).))).......(((((((.((((....))))..)))))))......... (-14.56 = -15.45 + 0.89)

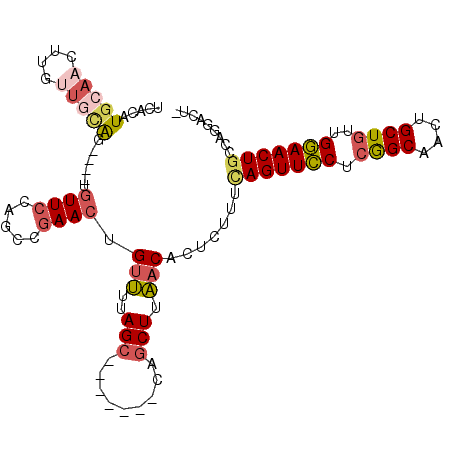
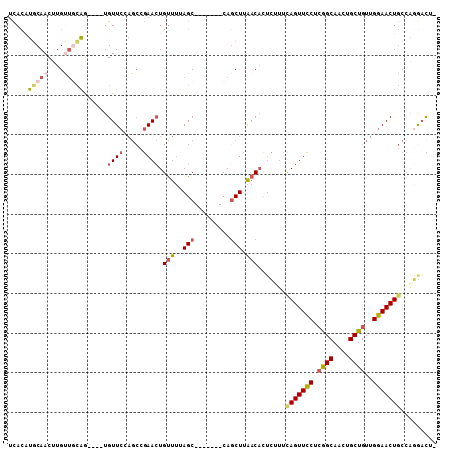
| Location | 9,019,632 – 9,019,726 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9019632 94 + 22407834 UUCAGUUCCUCGGCAAAUGCUGUUGGAACUGCCGGGACUACGCAAGGACUACGCUCCUUUGGAUCUUGAGAUUGCGUUAAAAUUACGUGUUCCA ..(((((((.((((....))))..)))))))..(((((.(((.((.....((((..(((........)))...)))).....)).)))))))). ( -31.30) >DroSec_CAF1 13606 94 + 1 UUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCGGGACGCCGCAAGGACUACGCUCCUUUGGAUCUUGAGAUUGCGUUAAAAUUACGUGUUCCA ..(((((((.((((....))))..)))))))..(((((((.(((((((......)))))..((((....))))))((.......))))))))). ( -32.60) >DroSim_CAF1 14639 94 + 1 UUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGACCGCCGCAAGGACUACGCUCCUUUGGAUCUUGAGAUUGCGUUAAAAUUACGUGUUCCA ..(((((((.((((....))))..)))))))(.(((...(((.(((((......)))))))).))).).((..((((.......))))..)).. ( -27.00) >DroEre_CAF1 12810 93 + 1 UUUAGUUCCGCGGCAACUGCUGUUGGAACUGCCACGAUUCCGCAAGGACUACGCUCCUGCGAAGCUUGGUAUAACGUGAAA-CUACGUGUUCCA ...(((((((((((....)))).)))))))((((.(..(((....)))((.(((....))).))).))))...(((((...-.)))))...... ( -31.20) >DroYak_CAF1 13375 94 + 1 UUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGGACUCCGCAAGGACUACGCUCCUUCGGAUCUUGAGAUAAUGUUAAAGUUACGUGUGCCA ..(((((((.((((....))))..))))))).(((((.((((.(((((......)))))))))))))).....((((.......))))...... ( -31.90) >DroAna_CAF1 18787 92 + 1 UUCAGUUCCAUGGUGACUGCUUUUGGAACUGCCAGAACUUCGCAAGGACUACGCUUCUAUUUGCCUCACUAUC--ACUAAAAUUACGUAGUCCC ..((((((((.(((....)))..))))))))..............((((((((.....((((...........--....))))..)))))))). ( -23.16) >consensus UUCAGUUCCUCGGCAACUGCUGUUGGAACUGCCAGGACUCCGCAAGGACUACGCUCCUUUGGAUCUUGAGAUUGCGUUAAAAUUACGUGUUCCA ..(((((((.((((....))))..)))))))..............(((.((((................................)))).))). (-17.85 = -17.77 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:37 2006