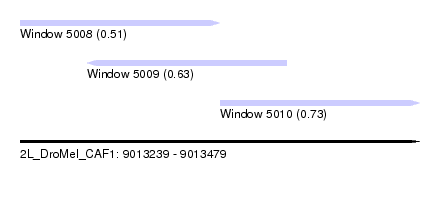
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,013,239 – 9,013,479 |
| Length | 240 |
| Max. P | 0.726813 |

| Location | 9,013,239 – 9,013,359 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -35.66 |
| Energy contribution | -36.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9013239 120 + 22407834 ACAUGCUUCAGCACCCGGAGGCAUGGAAACUUCAACAUAUGGAUGACCAUUUUGCCCAGGGUGAUGGCCACCUGCGAGAAGCGAUCGUGGUUCUCGCAAAUGUACGUCAGACCCAUCUCA .((((((((........))))))))..........((.((((....))))..))....((((((((..((..((((((((.((....)).))))))))..))..))))..))))...... ( -39.60) >DroSec_CAF1 7163 120 + 1 ACAUGCUUCAGCACCCGGAAGCAUGGAGAUUUCAUCAUUCGAAUGGUCAUUUUGCCCAGGGUGAUGGCCACCCCCGCGAAGCGAUCGUGGGUCUCGCAGAUGUACGAUAGACCCGUCUCA .((((((((........))))))))(((((............((((((.((((((...(((((.....)))))..)))))).))))))((((((((........)))..)))))))))). ( -46.50) >DroSim_CAF1 8154 120 + 1 ACACGCUUCAGCACCCGGAAGCAUGGAUACUUCAUCAUUCGGAUGGUCAUUUUGCCCAGGGUGGUGGCCACCCGAGCGAAGCGAUCGUGGGUCUCGCAGAUGUACGAUAGACCCGUCUUA ...((((((.((.....((((........)))).....((((.((((((((..(((...))))))))))).))))))))))))((((((.(((.....))).))))))(((....))).. ( -42.10) >consensus ACAUGCUUCAGCACCCGGAAGCAUGGAAACUUCAUCAUUCGGAUGGUCAUUUUGCCCAGGGUGAUGGCCACCCGCGCGAAGCGAUCGUGGGUCUCGCAGAUGUACGAUAGACCCGUCUCA .((((((((........)))))))).................((((((.((((((...(((((.....)))))..)))))).))))))((((((..(........)..))))))...... (-35.66 = -36.33 + 0.67)

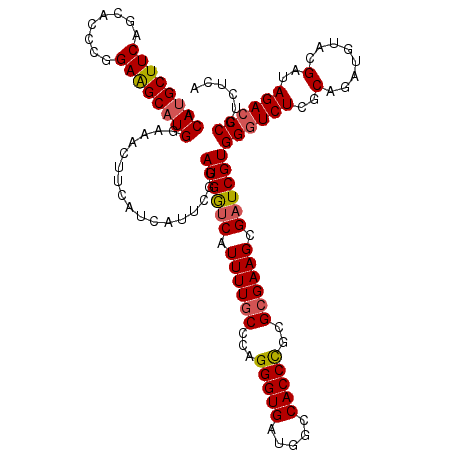
| Location | 9,013,279 – 9,013,399 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.47 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9013279 120 - 22407834 AGAUCGCGGCCACUUCAAUCCUUGAGAAGAUACUGCUCGAUGAGAUGGGUCUGACGUACAUUUGCGAGAACCACGAUCGCUUCUCGCAGGUGGCCAUCACCCUGGGCAAAAUGGUCAUCC .......(((((((((.........))))....((((((..(.((((((.....)...((((((((((((.(......).)))))))))))).))))).)..))))))...))))).... ( -38.70) >DroSec_CAF1 7203 120 - 1 AGUUUGCGGCCAUCAAAAUCAUUGAGAAGAUACUGCUCAAUGAGACGGGUCUAUCGUACAUCUGCGAGACCCACGAUCGCUUCGCGGGGGUGGCCAUCACCCUGGGCAAAAUGACCAUUC ..((((((((.(((....((((((((.((...)).))))))))...(((((..(((((....))))))))))..))).)))...(.((((((.....)))))).)))))).......... ( -43.00) >DroSim_CAF1 8194 120 - 1 AGUCUGCGGCCAUCAAAAUCAUUGAGAAGAUACUGCUCAAUAAGACGGGUCUAUCGUACAUCUGCGAGACCCACGAUCGCUUCGCUCGGGUGGCCACCACCCUGGGCAAAAUGACCAUCC .(((.(((..(.........((((((.((...)).)))))).....(((((..(((((....))))))))))..)..)))...((((((((((...)))))).)))).....)))..... ( -41.50) >consensus AGUUUGCGGCCAUCAAAAUCAUUGAGAAGAUACUGCUCAAUGAGACGGGUCUAUCGUACAUCUGCGAGACCCACGAUCGCUUCGCGCGGGUGGCCAUCACCCUGGGCAAAAUGACCAUCC .....(((((((((....((((((((.((...)).))))))))...((((((..((((....))))))))))................))))))).((.....))))............. (-32.35 = -32.47 + 0.12)

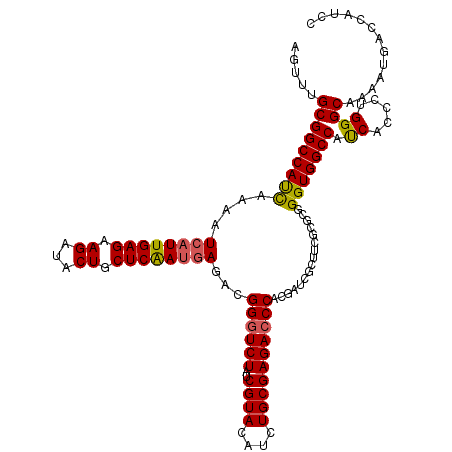
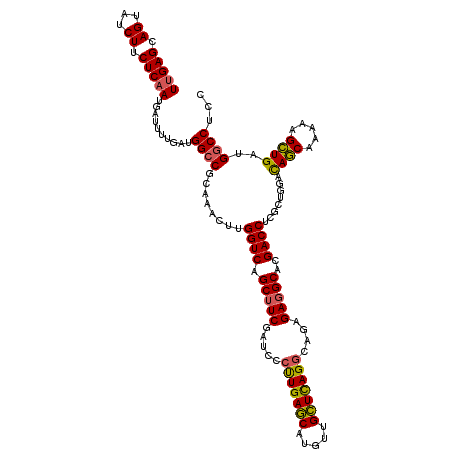
| Location | 9,013,359 – 9,013,479 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -31.98 |
| Energy contribution | -31.77 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9013359 120 + 22407834 UCGAGCAGUAUCUUCUCAAGGAUUGAAGUGGCCGCGAUCUUGGUCAGCUUCGAUCCCCUGACCAUAUUGGUCAGACAGUGAGGCACGACCUCGCUCCAUAUCAGGAAGAUGAGGACCUCC ..((((..((((((((...(((((((((((((((......)))))).))))))))).((((((.....))))))..(((((((.....)))))))........))))))))..)..))). ( -48.70) >DroSec_CAF1 7283 120 + 1 UUGAGCAGUAUCUUCUCAAUGAUUUUGAUGGCCGCAAACUUGGUCAGCAUCGAUCCCUUGAGCAUGUUGCUCAGGCAGAGAGGCACGACCUCGGUGGACAGCAAAAAGCUGAUGGCCUCC (((((((((((...(((((.((((.((.((((((......)))))).))..))))..))))).))))))))))).....(((((.(.((....)).).((((.....))))...))))). ( -40.20) >DroSim_CAF1 8274 120 + 1 UUGAGCAGUAUCUUCUCAAUGAUUUUGAUGGCCGCAGACUUGGUCAGCUUCGAUCCCUUGAGCAUGUUGCUUAGGCAGAGAGGCACGACCUCGCUGGACAGCAAAAAGCUGAUGGCCUCC (((((.((...)).))))).......((.((((((((.(((((((.(((((.....(((((((.....)))))))....)))))..))))..(((....)))...)))))).))))))). ( -40.60) >consensus UUGAGCAGUAUCUUCUCAAUGAUUUUGAUGGCCGCAAACUUGGUCAGCUUCGAUCCCUUGAGCAUGUUGCUCAGGCAGAGAGGCACGACCUCGCUGGACAGCAAAAAGCUGAUGGCCUCC (((((.((...)).)))))..........((((........((((.(((((.....(((((((.....)))))))....)))))..))))........((((.....))))..))))... (-31.98 = -31.77 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:35 2006