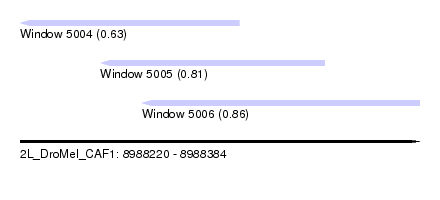
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,988,220 – 8,988,384 |
| Length | 164 |
| Max. P | 0.859776 |

| Location | 8,988,220 – 8,988,310 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.80 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -8.34 |
| Energy contribution | -7.82 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
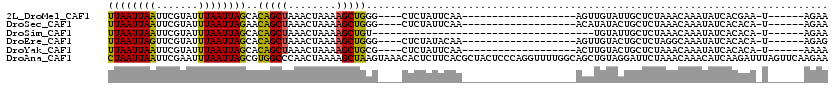
>2L_DroMel_CAF1 8988220 90 - 22407834 UUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG----CUCUAUUCAA-------------------AGUUGUAUUGCUCUAAACAAAUAUCACGAA-U------AGAA .(((((((.......)))))))((.(((((........))))).)----)(((((((..-------------------.((.(((((..........))))).)))))-)------))). ( -18.60) >DroSec_CAF1 32470 90 - 1 UUAAUUAAUUCGUAUUUAAUUAGAACAGCUAAACUAAAAGCUGGG----CUCUAUUCAA-------------------ACAUAUACUGCUCUAAACAAAUAUCACACA-U------AGAA ...........((((((..(((((.(((((........))))).(----(..(((....-------------------....)))..)))))))..))))))......-.------.... ( -12.90) >DroSim_CAF1 25909 76 - 1 UUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGU-------------------------------------UGUAUUGCUCUAAACAAAUAUCACACA-U------AGAA .(((((((.......)))))))((((((((........))))).-------------------------------------)))........................-.------.... ( -10.20) >DroEre_CAF1 55170 90 - 1 UUAAUUAGUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG----CUCUAUACAA-------------------AGUUGUACUGCUCUAGGCAAAUAUCACACA-U------AGAG .(((((((.......)))))))((.(((((........))))).)----)((((((((.-------------------...)))..(((.....)))..........)-)------))). ( -15.50) >DroYak_CAF1 51038 90 - 1 UUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGCG----CUCUAUUCAA-------------------ACUUGUACUGCUCUAAACAAAUAUCACACA-U------AAAA .(((((((.......)))))))((.(((((........))))).)----).........-------------------..............................-.------.... ( -11.90) >DroAna_CAF1 66804 120 - 1 CUAAUUAAUUCGAAUUUAAUUAGCGUGGCCCAACUAAAAGCUAAGUAAACACUCUUCACGCUACUCCCAGGUUUUGGCAGCUGUAGGAUUCUAAACAAACAUCAAGAUUUAGUUCAAGAA ..((((((.((...((((.(((((.(((.....)))...))))).))))...((((((.(((....((((...)))).))))).)))).................)).))))))...... ( -15.80) >consensus UUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG____CUCUAUUCAA___________________AGUUGUACUGCUCUAAACAAAUAUCACACA_U______AGAA ((((((((.......))))))))..(((((........)))))............................................................................. ( -8.34 = -7.82 + -0.52)
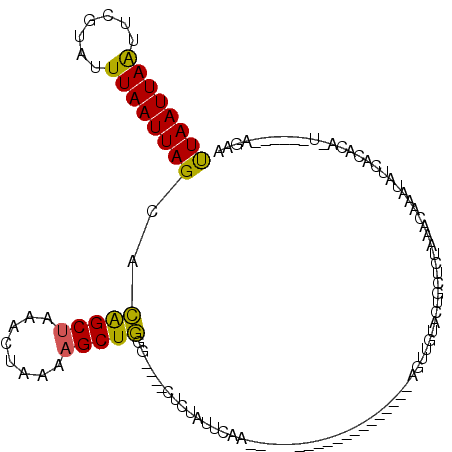
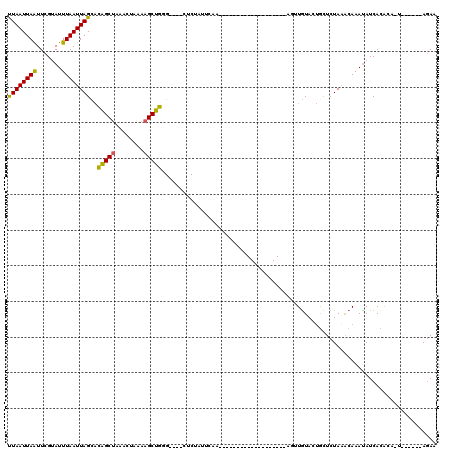
| Location | 8,988,253 – 8,988,345 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -13.85 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8988253 92 - 22407834 GUUAUGGUGAGAACAC-AUUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG----CUCUAUUCAA-------------------AG ((((..(((......)-))..)).))----..((((..((((((((((.......))))))))))(((((........))))).)----).))......-------------------.. ( -20.00) >DroSec_CAF1 32503 92 - 1 GUUAUGGUGAGAACAC-ACUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGAACAGCUAAACUAAAAGCUGGG----CUCUAUUCAA-------------------AC (((((((((......)-)))))).))----..((((....((((((((.......))))))))..(((((........))))).)----).))......-------------------.. ( -17.20) >DroSim_CAF1 25941 79 - 1 GUUAUGGUGAGAACAC-ACUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGU------------------------------------ (((((((((......)-)))))).))----.........(((((((((.......)))))))))((((((........))))))------------------------------------ ( -18.50) >DroEre_CAF1 55203 92 - 1 GUUAUGGUGAGAAUAA-ACUGUACAC----GCAGGCAUUGUUAAUUAGUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG----CUCUAUACAA-------------------AG ......((((((....-.((((....----))))((..((((((((((.......))))))))))(((((........))))).)----)))).)))..-------------------.. ( -20.70) >DroYak_CAF1 51071 96 - 1 GUUAUGGUGAGAACUC-ACUACACACGCACGCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGCG----CUCUAUUCAA-------------------AC ....((((((....))-))))...........((((..((((((((((.......))))))))))(((((........))))).)----).))......-------------------.. ( -21.10) >DroAna_CAF1 66844 116 - 1 UUUAUGGUGAGAAAAAAAGAGCACAC----GUAUGCAAUGCUAAUUAAUUCGAAUUUAAUUAGCGUGGCCCAACUAAAAGCUAAGUAAACACUCUUCACGCUACUCCCAGGUUUUGGCAG ....(((.(((.....(((((.....----....((.(((((((((((.......))))))))))).))...(((........))).....))))).......))))))........... ( -22.10) >consensus GUUAUGGUGAGAACAC_ACUGUACAC____GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAGCUGGG____CUCUAUUCAA___________________AG ..................((((........))))....((((((((((.......))))))))))(((((........)))))..................................... (-13.85 = -12.97 + -0.88)
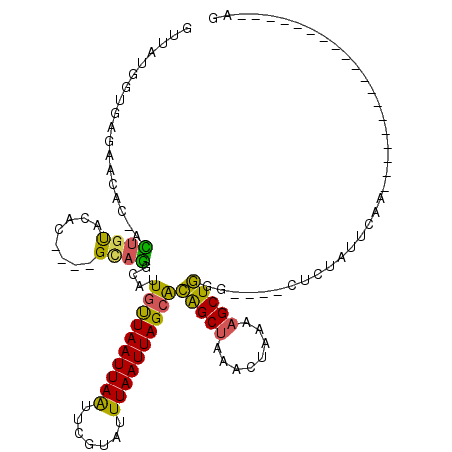
| Location | 8,988,270 – 8,988,384 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -14.08 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8988270 114 - 22407834 ACAAAAAAAAAAAAGC-GAAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAACAC-AUUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAG ..............((-(..((((.....))))......(((.(..(((......)-))..)))))----)).(((..((((((((((.......))))))))))..))).......... ( -22.30) >DroSec_CAF1 32520 114 - 1 ACAACAAAAAAAAAGC-GAAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAACAC-ACUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGAACAGCUAAACUAAAAG ..............((-(..((((.....))))......(((.((((((......)-)))))))))----)).(((....((((((((.......))))))))....))).......... ( -19.60) >DroSim_CAF1 25945 110 - 1 ACA----AAAAAAAGC-GAAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAACAC-ACUGUACAC----GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAG ...----.......((-(..((((.....))))......(((.((((((......)-)))))))))----)).(((..((((((((((.......))))))))))..))).......... ( -23.80) >DroEre_CAF1 55220 108 - 1 ACAA-------AAAGGGGCAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAAUAA-ACUGUACAC----GCAGGCAUUGUUAAUUAGUUCGUAUUUAAUUAGCACAGCUAAACUAAAAG ....-------....(..((((((((....(((((...)))))...))))......-))))..)..----...(((..((((((((((.......))))))))))..))).......... ( -20.40) >DroYak_CAF1 51088 111 - 1 ACCA-------AAAGC-GAAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAACUC-ACUACACACGCACGCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAG ....-------...((-(..((((.....))))......(((..((((((....))-)))).)))))).....(((..((((((((((.......))))))))))..))).......... ( -24.50) >DroAna_CAF1 66884 94 - 1 ACAU-------A---C-AAAGUUUUU-----------AGUUUUAUGGUGAGAAAAAAAGAGCACAC----GUAUGCAAUGCUAAUUAAUUCGAAUUUAAUUAGCGUGGCCCAACUAAAAG ....-------.---.-.....((((-----------((((.(((((((............))).)----))).((.(((((((((((.......))))))))))).))..)))))))). ( -20.80) >consensus ACAA_______AAAGC_GAAGUUCACAGAGAACAUAAAAUGUUAUGGUGAGAACAC_ACUGUACAC____GCAGGCAUUGUUAAUUAAUUCGUAUUUAAUUAGCACAGCUAAACUAAAAG .....................(((((....((((.....))))...))))).......((((........))))((..((((((((((.......))))))))))..))........... (-14.08 = -14.00 + -0.08)

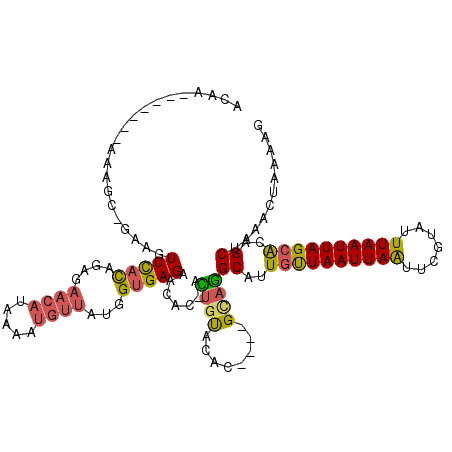
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:31 2006