
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,984,884 – 8,984,982 |
| Length | 98 |
| Max. P | 0.995601 |

| Location | 8,984,884 – 8,984,982 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -31.48 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
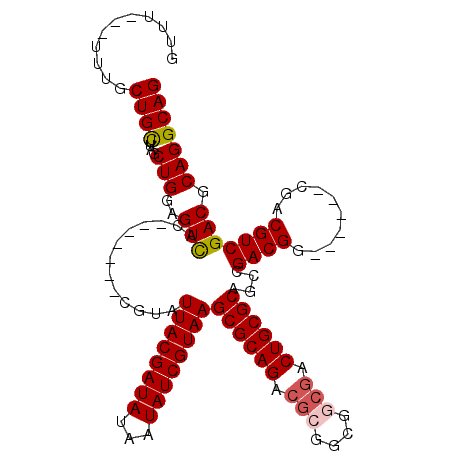
>2L_DroMel_CAF1 8984884 98 + 22407834 GUU-----UUGCUGUUUUCCUGGAGUUAC-------CGUAUUACGAUAUAAUAUCGUAAGCGCAGACGCGGCGGCGACUGCGCAGCCGACGG------UGACGUCGACGCAGGCAG ...-----...((((((.(.(.(((((((-------(((.((((((((...))))))))((((((.(((....))).)))))).....))))------)))).)).).).)))))) ( -40.40) >DroSec_CAF1 29003 101 + 1 GUUUU--UUUGCUGCUUAGCUGGAGUCAC-------CGUAUUACGAUAUAAUAUCGUAAGCGCAGACGCGGCGGCGACUGCGCUGGCGACGG------CGACGUCGACGCAGGCAG .....--....((((((.(((.(((((.(-------(((..(((((((...)))))))(((((((.(((....))).)))))))....))))------.))).)).).)))))))) ( -43.10) >DroSim_CAF1 22416 103 + 1 GUUUUUUUUUGCUGCUUAGCUGGAGUCAC-------CGUAUUACGAUAUAAUAUCGUAAGCGCAGACGCGGCGCCGACUGCGCAGCCGACGG------CGACGUCGACGCAGGCAG ...........((((((.(((.(((((.(-------(((.((((((((...))))))))((((((...((....)).)))))).....))))------.))).)).).)))))))) ( -40.50) >DroYak_CAF1 47376 107 + 1 AUUU---UGUGCUGCUUUGCUGGAGUCACCGUAUAUCGUAUUACGAUAUAAUAUCGUAAGCGCAGACGCGG------CUGCGCAGCCGACGGCGGCGGCGACGUCGACGCAGGCAG ....---(((.((((.(((.((..(((.((((...(((..((((((((...))))))))((((((......------))))))...)))..)))).)))..)).))).))))))). ( -42.30) >consensus GUUU___UUUGCUGCUUAGCUGGAGUCAC_______CGUAUUACGAUAUAAUAUCGUAAGCGCAGACGCGGCGGCGACUGCGCAGCCGACGG______CGACGUCGACGCAGGCAG ...........((((....(((..(((.............((((((((...))))))))((((((.(((....))).))))))....((((..........))))))).))))))) (-31.48 = -32.10 + 0.62)

| Location | 8,984,884 – 8,984,982 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -31.05 |
| Energy contribution | -32.05 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8984884 98 - 22407834 CUGCCUGCGUCGACGUCA------CCGUCGGCUGCGCAGUCGCCGCCGCGUCUGCGCUUACGAUAUUAUAUCGUAAUACG-------GUAACUCCAGGAAAACAGCAA-----AAC ...((((.(((((((...------.))))))).((((((.(((....))).))))))((((((((...))))))))....-------.......))))..........-----... ( -36.80) >DroSec_CAF1 29003 101 - 1 CUGCCUGCGUCGACGUCG------CCGUCGCCAGCGCAGUCGCCGCCGCGUCUGCGCUUACGAUAUUAUAUCGUAAUACG-------GUGACUCCAGCUAAGCAGCAAA--AAAAC ((((..((...((.((((------((((.....((((((.(((....))).))))))((((((((...)))))))).)))-------)))))))..))...))))....--..... ( -43.10) >DroSim_CAF1 22416 103 - 1 CUGCCUGCGUCGACGUCG------CCGUCGGCUGCGCAGUCGGCGCCGCGUCUGCGCUUACGAUAUUAUAUCGUAAUACG-------GUGACUCCAGCUAAGCAGCAAAAAAAAAC ((((..((...((.((((------((((.....((((((.((......)).))))))((((((((...)))))))).)))-------)))))))..))...))))........... ( -40.20) >DroYak_CAF1 47376 107 - 1 CUGCCUGCGUCGACGUCGCCGCCGCCGUCGGCUGCGCAG------CCGCGUCUGCGCUUACGAUAUUAUAUCGUAAUACGAUAUACGGUGACUCCAGCAAAGCAGCACA---AAAU ((((.(((...((.(((((((.....((((...((((((------......))))))...))))..(((((((.....))))))))))))))))..)))..))))....---.... ( -41.50) >consensus CUGCCUGCGUCGACGUCG______CCGUCGGCUGCGCAGUCGCCGCCGCGUCUGCGCUUACGAUAUUAUAUCGUAAUACG_______GUGACUCCAGCAAAGCAGCAAA___AAAC .(((.((((((((((..........))))))).((((((.((......)).))))))((((((((...)))))))).........................))))))......... (-31.05 = -32.05 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:29 2006