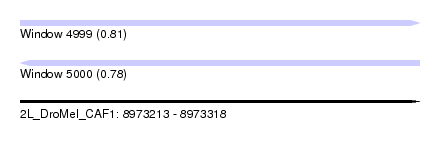
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,973,213 – 8,973,318 |
| Length | 105 |
| Max. P | 0.812191 |

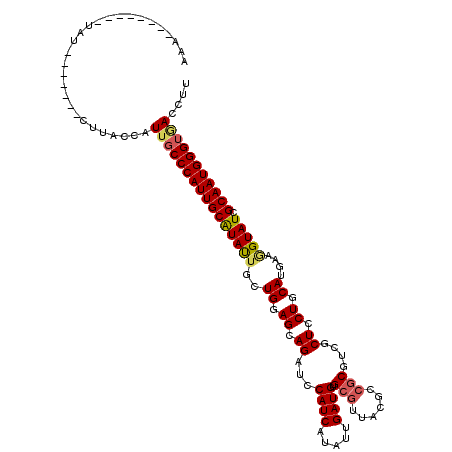
| Location | 8,973,213 – 8,973,318 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8973213 105 + 22407834 AGA--------UAU-------CUUACCAUUGCCCAUUGCAUAUUGCUGGAGGAGAUCCAUCAUAUUGAUGUGCGUUACACCGCGUCGCUCCUGCAUGAAGGUAUCGCAAUGGGUGACCUU ...--------...-------.......(..(((((((((((((..((.(((((...((((.....)))).(((......)))....))))).))....))))).))))))))..).... ( -35.20) >DroEre_CAF1 36752 110 + 1 AUAUGCU---AUAU-------CUUACCAUUGCCCAUUGCAUACUGCUGGAGGAGAUCCAUCAUAUUGAUGUGCGUUACGCCGCGUCGCUCCUGCAUGAAGGUAUCGCAAUGGGUGACCUU .......---....-------.......(..(((((((((((((..((.(((((...((((.....)))).(((......)))....))))).))....))))).))))))))..).... ( -37.60) >DroWil_CAF1 92361 107 + 1 AAAUC------CAA-------CUUACCAUUGCCCAUUGCGUAUUGUUGCAGCAGAUCCAUCAUAUUGAUGUGGAUGACUCCGCGUCUCUCCUGCAUAAAGGUAUCGCAAUGGGUAACUUU .....------...-------.......((((((((((((((((..(((((.(((...........((((((((....))))))))))).)))))....)))).)))))))))))).... ( -39.40) >DroYak_CAF1 35372 120 + 1 AAAUACUCGUAUAUACUCGUACUUACCAUUGCCCAUUGCAUAUUGCUGGAGGAGAUCCAUCAUAUUGAUGUGCGUUACGCCGCGUCGCUCCUGCAUGAAGGUAUCGCAAUGGGUGACCUU ........((((......))))......(..(((((((((((((..((.(((((...((((.....)))).(((......)))....))))).))....))))).))))))))..).... ( -36.50) >DroMoj_CAF1 33055 101 + 1 ------------CA-------CUUACCGUUUCCCAUUGCGUAUUGCUGCAGCAGAUCCAUCAUAUUGAUGUGCGUAACGCCGCGUCGCUCCUGCAUGAAUGUAUCGCAAUGGGUUACCUU ------------..-------......((..(((((((((...(((..((((((...((((.....)))).(((......))).......)))).))...))).)))))))))..))... ( -33.20) >DroAna_CAF1 51704 105 + 1 UAA--------UAU-------CUUACCAUUGCCCAUUGCAUAUUGCUGAAGCAGAUCCAUCAUAUUGAUGUGCGUCACUCCGCGUCGCUCCUGCAUAAAAGUAUCGCAAUGGGUGACUUU ...--------...-------.......(..(((((((((((((......((((...((((.....)))).(((.(.......).)))..)))).....))))).))))))))..).... ( -30.80) >consensus AAA________UAU_______CUUACCAUUGCCCAUUGCAUAUUGCUGGAGCAGAUCCAUCAUAUUGAUGUGCGUUACGCCGCGUCGCUCCUGCAUGAAGGUAUCGCAAUGGGUGACCUU ............................((((((((((((((((..((.((.((...((((.....)))).(((......)))....)).)).))....))))).))))))))))).... (-25.38 = -25.63 + 0.25)


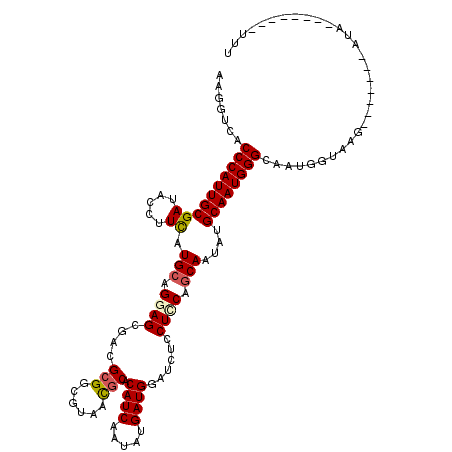
| Location | 8,973,213 – 8,973,318 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -26.43 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8973213 105 - 22407834 AAGGUCACCCAUUGCGAUACCUUCAUGCAGGAGCGACGCGGUGUAACGCACAUCAAUAUGAUGGAUCUCCUCCAGCAAUAUGCAAUGGGCAAUGGUAAG-------AUA--------UCU .......((((((((((.....)).(((.((((.((.(((......))).((((.....))))....)))))).)))....))))))))..........-------...--------... ( -32.70) >DroEre_CAF1 36752 110 - 1 AAGGUCACCCAUUGCGAUACCUUCAUGCAGGAGCGACGCGGCGUAACGCACAUCAAUAUGAUGGAUCUCCUCCAGCAGUAUGCAAUGGGCAAUGGUAAG-------AUAU---AGCAUAU .......((((((((.((((.....(((.((((.((.(((......))).((((.....))))....)))))).)))))))))))))))..(((.((..-------...)---).))).. ( -34.20) >DroWil_CAF1 92361 107 - 1 AAAGUUACCCAUUGCGAUACCUUUAUGCAGGAGAGACGCGGAGUCAUCCACAUCAAUAUGAUGGAUCUGCUGCAACAAUACGCAAUGGGCAAUGGUAAG-------UUG------GAUUU .......(((((((((.........(((((.((((((.....))).(((((((....))).))))))).)))))......)))))))))((((.....)-------)))------..... ( -33.96) >DroYak_CAF1 35372 120 - 1 AAGGUCACCCAUUGCGAUACCUUCAUGCAGGAGCGACGCGGCGUAACGCACAUCAAUAUGAUGGAUCUCCUCCAGCAAUAUGCAAUGGGCAAUGGUAAGUACGAGUAUAUACGAGUAUUU .......((((((((((.....)).(((.((((.((.(((......))).((((.....))))....)))))).)))....)))))))).......((((((..((....))..)))))) ( -36.60) >DroMoj_CAF1 33055 101 - 1 AAGGUAACCCAUUGCGAUACAUUCAUGCAGGAGCGACGCGGCGUUACGCACAUCAAUAUGAUGGAUCUGCUGCAGCAAUACGCAAUGGGAAACGGUAAG-------UG------------ ...((..(((((((((.........(((((.((....(((......))).((((.....))))...)).)))))......)))))))))..))......-------..------------ ( -33.46) >DroAna_CAF1 51704 105 - 1 AAAGUCACCCAUUGCGAUACUUUUAUGCAGGAGCGACGCGGAGUGACGCACAUCAAUAUGAUGGAUCUGCUUCAGCAAUAUGCAAUGGGCAAUGGUAAG-------AUA--------UUA .......((((((((((.....)).(((.((((((..(((......))).((((.....))))....)))))).)))....))))))))..........-------...--------... ( -30.40) >consensus AAGGUCACCCAUUGCGAUACCUUCAUGCAGGAGCGACGCGGCGUAACGCACAUCAAUAUGAUGGAUCUCCUCCAGCAAUAUGCAAUGGGCAAUGGUAAG_______AUA________UUU .......((((((((((.....)).(((.((((....(((......))).((((.....))))......)))).)))....))))))))............................... (-26.43 = -26.65 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:26 2006