
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
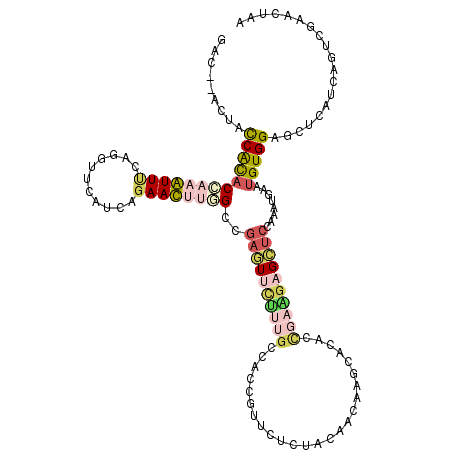
| Location | 8,964,390 – 8,964,508 |
| Length | 118 |
| Max. P | 0.835851 |

| Location | 8,964,390 – 8,964,508 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.65 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -11.44 |
| Energy contribution | -12.48 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
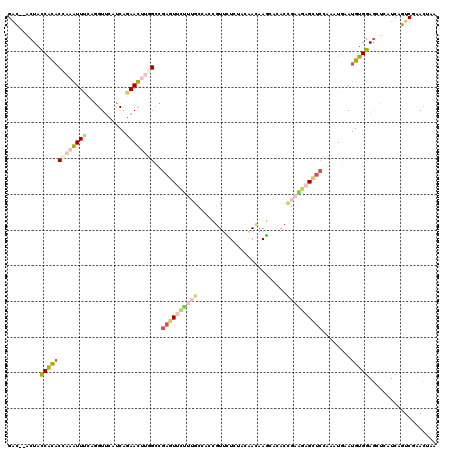
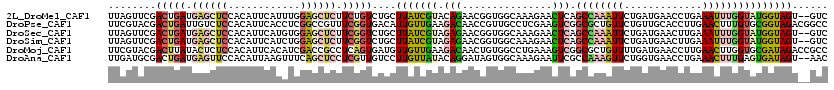
>2L_DroMel_CAF1 8964390 118 + 22407834 GAC--ACUACCAUACCAAAUUUCAGGUUCAUCAGAAUUUGGCUGAGUUCUUUGCCACCGUUCUGUACGAUAAGCAGACAGAAGAGCUCCAAAUGAAUGUGGAGCUCAUCAGUCGAACUAA ...--...................(((((..((((((.((((.((....)).))))..))))))...........(((.((.((((((((........)))))))).)).)))))))).. ( -35.20) >DroPse_CAF1 12420 120 + 1 GGCCGUCUACCGCACAAAGUUCAAGGUGCAACAGAACAGCGCCGACUUCGAGGCAACGGUUGUCUUCAACAAUGUCACCGAAACGGCCGAGGUGAAUGUGGAGACAAUCAGUCGUACGAA ((((.(((...((((..........))))...)))...).)))((((....(....)(((((((((((.((...(((((...........))))).)))))))))))))))))....... ( -38.90) >DroSec_CAF1 8755 118 + 1 GAC--ACUACCAUACCAAAUUUCAAGUUCAUCAGAAUUUGGCUGAGUUCUUUGCCACCGUUCUCUACGAUAAGCAGACCGAAGAGCUCCACAUGAAUGUGGAGCUCAUCAGUCGAACUAA ...--................((((((((....))))))))...(((((..(((...(((.....)))....)))(((.((.((((((((((....)))))))))).)).)))))))).. ( -38.30) >DroSim_CAF1 2435 118 + 1 GAC--ACUACCAUACCAAAUUUCAAGUUCAUCAGAAUUUGGCUGAGUUCUUUGCCACCGUUCUCUACGAUAAGCAGACCGAAGAGCUCCAGAUGAAUGUGGAGCUCAUCAGUCGAACUAA ...--................((((((((....))))))))...(((((..(((...(((.....)))....)))(((.((.((((((((.(....).)))))))).)).)))))))).. ( -34.40) >DroMoj_CAF1 11638 120 + 1 GGCGGUCUAUCGCACCAAGUUCAAGGUUCAUCAAAACAGCGCCGACUUUCAGGCCACAGUUGUCUUCAACAACAUCACUGAGGCGGUCGAUGUGAAUGUGGAGAGUAUAAGUCGUACGAA ((((((.....))(((........)))............))))(((((.....(((((((((....)))).(((((((((...)))).)))))...))))).......)))))....... ( -30.00) >DroAna_CAF1 34197 118 + 1 GUU--ACUAUCACUCAAAGUUUCAGGUUCACCAGAACUUUGGCGAAUUCUUUGCCACUAUCCUGUAUAACAAGGACAACGAGGAGCUGAAACUUAAUGUGGAACUCAUCAGUCGCAUCAA ((.--(((..(((...(((((((((.(((..........((((((.....))))))...((((........))))......))).)))))))))...)))((.....))))).))..... ( -30.20) >consensus GAC__ACUACCACACCAAAUUUCAGGUUCAUCAGAACUUGGCCGAGUUCUUUGCCACCGUUCUCUACAACAAGCACACCGAAGAGCUCCAAAUGAAUGUGGAGCUCAUCAGUCGAACUAA .........(((((((((((((...........))))))))..((((((((((.........................))))))))))........)))))................... (-11.44 = -12.48 + 1.03)

| Location | 8,964,390 – 8,964,508 |
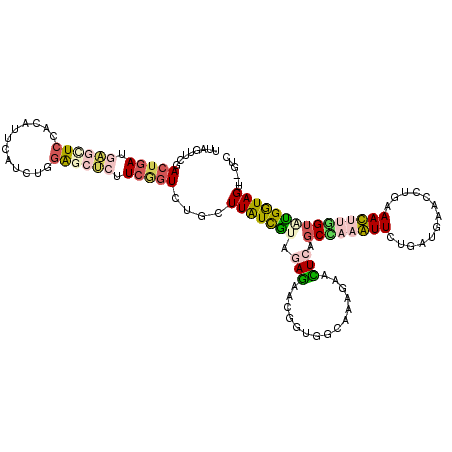
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.65 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.71 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8964390 118 - 22407834 UUAGUUCGACUGAUGAGCUCCACAUUCAUUUGGAGCUCUUCUGUCUGCUUAUCGUACAGAACGGUGGCAAAGAACUCAGCCAAAUUCUGAUGAACCUGAAAUUUGGUAUGGUAGU--GUC ..((((((((.((.((((((((........)))))))).)).)))((((.(((((.....)))))))))..)))))..(((((((((.(......).).))))))))........--... ( -38.20) >DroPse_CAF1 12420 120 - 1 UUCGUACGACUGAUUGUCUCCACAUUCACCUCGGCCGUUUCGGUGACAUUGUUGAAGACAACCGUUGCCUCGAAGUCGGCGCUGUUCUGUUGCACCUUGAACUUUGUGCGGUAGACGGCC ......(((.(((.(((....))).)))..)))((((((((((..((...((((....)))).))..))..)))).))))(((((.(((..((((..........))))..)))))))). ( -38.20) >DroSec_CAF1 8755 118 - 1 UUAGUUCGACUGAUGAGCUCCACAUUCAUGUGGAGCUCUUCGGUCUGCUUAUCGUAGAGAACGGUGGCAAAGAACUCAGCCAAAUUCUGAUGAACUUGAAAUUUGGUAUGGUAGU--GUC ..(((((((((((.((((((((((....)))))))))).))))))((((.(((((.....)))))))))..)))))..((((((((..((.....))..))))))))........--... ( -47.40) >DroSim_CAF1 2435 118 - 1 UUAGUUCGACUGAUGAGCUCCACAUUCAUCUGGAGCUCUUCGGUCUGCUUAUCGUAGAGAACGGUGGCAAAGAACUCAGCCAAAUUCUGAUGAACUUGAAAUUUGGUAUGGUAGU--GUC ..(((((((((((.((((((((........)))))))).))))))((((.(((((.....)))))))))..)))))..((((((((..((.....))..))))))))........--... ( -43.00) >DroMoj_CAF1 11638 120 - 1 UUCGUACGACUUAUACUCUCCACAUUCACAUCGACCGCCUCAGUGAUGUUGUUGAAGACAACUGUGGCCUGAAAGUCGGCGCUGUUUUGAUGAACCUUGAACUUGGUGCGAUAGACCGCC ...((.((((((....((.(((((.((((.............)))).((((((...)))))))))))...))))))))))((.((((...((.(((........))).))..)))).)). ( -26.12) >DroAna_CAF1 34197 118 - 1 UUGAUGCGACUGAUGAGUUCCACAUUAAGUUUCAGCUCCUCGUUGUCCUUGUUAUACAGGAUAGUGGCAAAGAAUUCGCCAAAGUUCUGGUGAACCUGAAACUUUGAGUGAUAGU--AAC ....(((.(((((.(((((..((.....))...))))).))...(((((.(.....))))))))).)))......((((((((((((.((....)).).))))))).))))....--... ( -30.90) >consensus UUAGUUCGACUGAUGAGCUCCACAUUCAUCUGGAGCUCUUCGGUCUGCUUAUCGUAGAGAACGGUGGCAAAGAACUCAGCCAAAUUCUGAUGAACCUGAAACUUGGUAUGGUAGU__GUC ........(((((.((((((............)))))).)))))....(((((((.(((...............))).((((((((.............)))))))))))))))...... (-12.32 = -12.71 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:23 2006