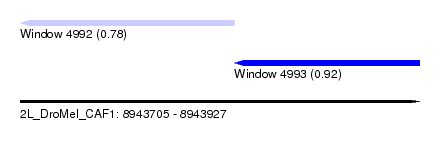
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,943,705 – 8,943,927 |
| Length | 222 |
| Max. P | 0.920424 |

| Location | 8,943,705 – 8,943,824 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.19 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8943705 119 - 22407834 UACCACUUCCAUAAAACAACUUUAUGUCCUCGGUCCAUUUCUUCACAUCCCGAAAAGGUUUCGU-UUGGGCUCCACUGCGGAGCUCUUCCCUUUUGUACAAAUACCAAAACUAGUUAUAA .(((.....((((((.....))))))...((((................))))...)))...((-(((((((((.....))))))).........(((....)))..))))......... ( -19.29) >DroSec_CAF1 154201 118 - 1 UACCACUUCCAUAAACCAACUUUAUGUCCUCGGUCCAUUUCUUCACAUCCCGAA-AGGUUUCGU-UUGGACUCCACUACGGAGCUCUUUCCUUUUGUACAAAUACCAAAACUAGGUAUAA ((((.....((((((.....)))))).....(((..((((....(((....(((-(((((((((-.(((...)))..))))))).)))))....)))..))))))).......))))... ( -23.40) >DroSim_CAF1 153675 118 - 1 UACCACUUCCAUAAACCAACUUUAUGUCCUCGGUCCAUUUCUUCACAUCCCGAA-AGGUUUCGU-UUGGGCUCCACUGCGGAGCUCUUUCCUUUUGUACAAAUACCAAAACUAGGUAUAA .(((.....((((((.....)))))).....)))................((((-(((......-..(((((((.....)))))))...))))))).....(((((.......))))).. ( -25.50) >DroYak_CAF1 158879 119 - 1 UACCACUUCCAUUAAUCAACUUUAUGUCGUCGGUCCAUUUAUCCACAUCCCGAA-AGGUUUCGUUUUGGGCUCUAUUGUAGAGCUCGUUCCUUUUGUACAAAUACCAAAACUAGGUAUAA ((((........................((.((.........))))...((...-.))....(((((((((((((...))))))..((.........)).....)))))))..))))... ( -22.40) >consensus UACCACUUCCAUAAACCAACUUUAUGUCCUCGGUCCAUUUCUUCACAUCCCGAA_AGGUUUCGU_UUGGGCUCCACUGCGGAGCUCUUUCCUUUUGUACAAAUACCAAAACUAGGUAUAA .(((.....((((((.....))))))...((((................))))...)))........(((((((.....)))))))...............(((((.......))))).. (-18.82 = -19.19 + 0.37)

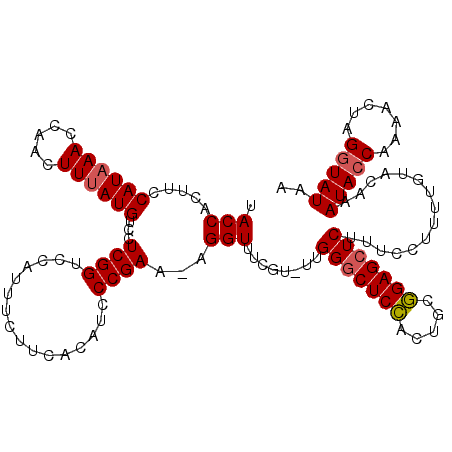
| Location | 8,943,824 – 8,943,927 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8943824 103 - 22407834 CACACGGGUAGACACCU-AAUGUACAUAUAGCUUUUGUAUAUAUCAAUAGAUAAUUGUGUAAUCCAACACGAUGAAUCGCUACAAGCUAUCCUAGCAAUGCCAU .....((((((.(....-.(((((((.........))))))).......(((.(((((((......)))))))..))))))))..((((...))))....)).. ( -21.60) >DroSec_CAF1 154319 103 - 1 CACACGGGUAGACCCGU-AAUGUACAUAUAGCUUUUGUAUAUAUCAAUAGAUAAUUGUGUAAUCCAACACGAUGAAUCGCUACAAGCUAUCCUAGCAAUGCCAU ...(((((....)))))-..(((....((((((..((((..((((....))))(((((((......))))))).......))))))))))....)))....... ( -24.10) >DroSim_CAF1 153793 103 - 1 CACACGGGUAGACUCCU-AAUGUACAUAUAGCUUUUGUAUAUAUCAAUAGAUAAUUGUGUAAUCCAACACGAUGAAUCGCUACAAGCUAUCCUAGCAAUGCCAU .....((((((......-.(((((((.........))))))).......(((.(((((((......)))))))..))).))))..((((...))))....)).. ( -21.30) >DroYak_CAF1 158998 104 - 1 CACACGGGUAGACUCUUCAAUGUACAUAUAGCUUUUGUAUAUAUGAAUAGAUAAUUGUGUAAUCCUACAUGAUAAAUCGCUGCAAGCUAUCCUAGCAAUGCCAU .....((((((.((.(((((((((((.........))))))).)))).))...((..((((....))))..))......))))..((((...))))....)).. ( -21.60) >consensus CACACGGGUAGACUCCU_AAUGUACAUAUAGCUUUUGUAUAUAUCAAUAGAUAAUUGUGUAAUCCAACACGAUGAAUCGCUACAAGCUAUCCUAGCAAUGCCAU .....((((((........(((((((.........))))))).......(((.(((((((......)))))))..))).))))..((((...))))....)).. (-20.70 = -20.32 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:19 2006