| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,937,544 – 8,937,649 |
| Length | 105 |
| Max. P | 0.836775 |

| Location | 8,937,544 – 8,937,649 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
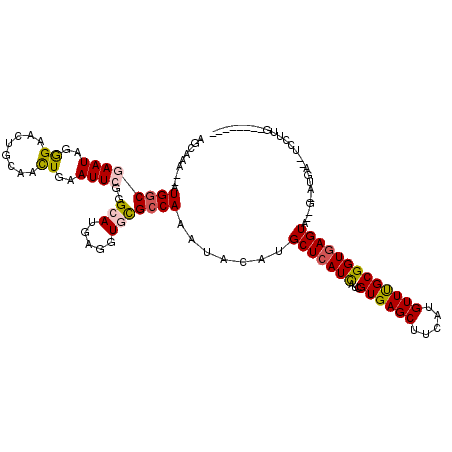
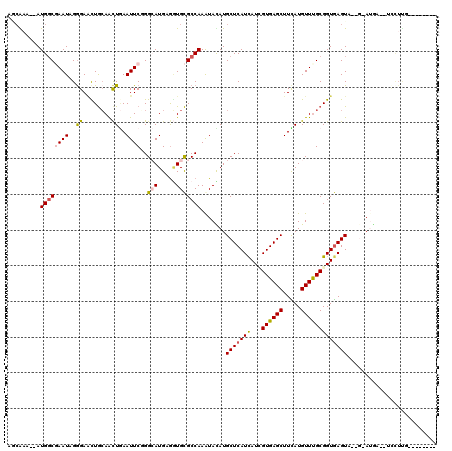
>2L_DroMel_CAF1 8937544 105 + 22407834 AGCAAA--AUGUCGAAUAGGGAACUGCAACUGAAUUCGGGCAUGAAGUGCGCCAAGUACAUGCUCAUCAUCGUGAGCUUCAUGUUUGCGGUGAGUA--G-AUGA--UCCUUG-------- ......--........((((((.((((..........((((((...((((.....)))))))))).((((((..(((.....)))..)))))))))--)-....--))))))-------- ( -32.70) >DroVir_CAF1 122180 113 + 1 AGCAAA--AUGGCGAAUCGGGAGCUGGAAUUAAAUUCCGGAAUGCGCUGCGCCAAAUAUAUGCUCAUCAUAGUGAGCUUUAUGUUCGCGGUGAGUUUUCCAAAAA----AAA-AAAAAAA ......--.(((((...((.(..(((((((...)))))))..).))...))))).......(((((((...((((((.....)))))))))))))..........----...-....... ( -33.00) >DroSec_CAF1 148055 105 + 1 AGCGAA--AUGGCGAAUAGGGAACUGCAGCUGAAUUCGGGCAUGAGGUGCGCCAAGUACAUGCUCAUCAUCGUGAGCUUCAUGUUUGCGGUGAGUA--G-AUGA--UCCUUG-------- .((...--...))...((((((.((((..........((((((...((((.....)))))))))).((((((..(((.....)))..)))))))))--)-....--))))))-------- ( -34.20) >DroSim_CAF1 147062 102 + 1 ---GAA--AUGGCUAAUAGAGAACUGCAGCUGAAUUCGGGCAUGAGGUGCGCCAAGUACAUGCUCAUCAUCGUGAGCUUCAUGUUUGCGGUGAGUA--G-AUGA--UCCUUG-------- ---...--...............((((..........((((((...((((.....)))))))))).((((((..(((.....)))..)))))))))--)-....--......-------- ( -29.10) >DroWil_CAF1 212942 117 + 1 AACGAAAUAUGGCUAAUCGGGAUUUACAACUGAAUUCCGGCAUGCGAUGUGCCAAAUAUAUGCUCAUCAUAGUGAGCUUUAUGUUUGCGGUAAGUUUUCAAAGAGUUUUUCA-AAACA-- ...(((((((.((.....((((((((....))))))))(((((.....)))))(((((((.((((((....))))))..))))))))).)))...)))).....((((....-)))).-- ( -31.30) >DroAna_CAF1 149604 105 + 1 UUCACA--AUGGCGAAUCGGGAAUUGCAGCUCAAUUCUGGAAUGCGGUGCGCCAAAUAUAUGCUCAUUAUUGUGAGCUUUAUGUUUGCGGUGAGUU--G-AU-----CCUUCAGG----- (((((.--((.(((..((.(((((((.....))))))).)).))).)).(((.(((((((.((((((....))))))..)))))))))))))))((--(-(.-----...)))).----- ( -34.00) >consensus AGCAAA__AUGGCGAAUAGGGAACUGCAACUGAAUUCGGGCAUGAGGUGCGCCAAAUACAUGCUCAUCAUCGUGAGCUUCAUGUUUGCGGUGAGUA__G_AUGA__UCCUUG________ .........((((((((..((........))..))))..(((.....))))))).......(((((((...((((((.....)))))))))))))......................... (-16.33 = -16.63 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:16 2006