| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
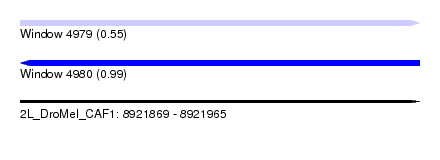
| Location | 8,921,869 – 8,921,965 |
| Length | 96 |
| Max. P | 0.988224 |

| Location | 8,921,869 – 8,921,965 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8921869 96 + 22407834 ACCGGCGGGUUAAAGUUUUCAUUUGGAGCAUAUUAUGCAUUUUAGCACUAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((.(((....)))((((...)))))))))))).)).........(((....((((((((...))))))))...))))))... ( -20.90) >DroSec_CAF1 132068 96 + 1 ACCGGCGGGUUAAAGUUUUCAUUUGGAGCAUAUUAUGCAUUUUAGCACUAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((.(((....)))((((...)))))))))))).)).........(((....((((((((...))))))))...))))))... ( -20.90) >DroSim_CAF1 131075 96 + 1 ACCGGCGGGUUAAAGUUUUCAUUUGGAGCAUUUUAUGCAUUUUAGCACUAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((.(((....)))((((...)))))))))))).)).........(((....((((((((...))))))))...))))))... ( -20.90) >DroEre_CAF1 139762 96 + 1 ACCGGCGGGUUAAAGUUUUCAUUUGGCGCACAUUAUGCAUUUUAGCACCAUAAAUCAUCAAUGCAUUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((...(((.((.....))..))).)))))))).)).........(((.....(((((((...)))))))....))))))... ( -21.90) >DroYak_CAF1 136347 96 + 1 ACCGGCGGGUUAAAGUUUUCAUUUGGAGCACAUUAUGCAUUUUAGCACUAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((.(((....)))(((.....))))))))))).)).........(((....((((((((...))))))))...))))))... ( -20.50) >DroAna_CAF1 134401 94 + 1 -CCGGCGGGUUA-AGUUUUCAACUCGAGUAUAUUAUACAUUUUAGCACCAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUGGGCCAGU -..(((((((((-((...((.....))((((...))))..)))))).))........((((....((((((((...))))))))..)))))))... ( -19.40) >consensus ACCGGCGGGUUAAAGUUUUCAUUUGGAGCAUAUUAUGCAUUUUAGCACUAUAAAUCAUCAAUGCAAUCGAUUGCCACAAUCGAUCUUUUGGCCAGU ...(((((((((((((.(((....)))(((.....))))))))))).)).........(((....((((((((...))))))))...))))))... (-18.93 = -19.27 + 0.33)


| Location | 8,921,869 – 8,921,965 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.11 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8921869 96 - 22407834 ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUAGUGCUAAAAUGCAUAAUAUGCUCCAAAUGAAAACUUUAACCCGCCGGU ((((((.....(((((((((...))))))))).((((.......(((.((((......)))).))).......)))).............)))))) ( -24.04) >DroSec_CAF1 132068 96 - 1 ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUAGUGCUAAAAUGCAUAAUAUGCUCCAAAUGAAAACUUUAACCCGCCGGU ((((((.....(((((((((...))))))))).((((.......(((.((((......)))).))).......)))).............)))))) ( -24.04) >DroSim_CAF1 131075 96 - 1 ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUAGUGCUAAAAUGCAUAAAAUGCUCCAAAUGAAAACUUUAACCCGCCGGU ((((((.....(((((((((...)))))))))(((((...........((((......))))..))))).....................)))))) ( -23.02) >DroEre_CAF1 139762 96 - 1 ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAAUGCAUUGAUGAUUUAUGGUGCUAAAAUGCAUAAUGUGCGCCAAAUGAAAACUUUAACCCGCCGGU ((((((..((((.(((((((...)))))))................((((((...............))))))........)))).....)))))) ( -22.96) >DroYak_CAF1 136347 96 - 1 ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUAGUGCUAAAAUGCAUAAUGUGCUCCAAAUGAAAACUUUAACCCGCCGGU ((((((.....(((((((((...))))))))).((((.......(((.((((......)))).))).......)))).............)))))) ( -23.74) >DroAna_CAF1 134401 94 - 1 ACUGGCCCAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUGGUGCUAAAAUGUAUAAUAUACUCGAGUUGAAAACU-UAACCCGCCGG- .(((((.(((.(((((((((...)))))))))...)))..((..(((.((((......)))).)))...))(.(((((....)-)))).))))))- ( -21.50) >consensus ACUGGCCAAAAGAUCGAUUGUGGCAAUCGAUUGCAUUGAUGAUUUAUAGUGCUAAAAUGCAUAAUAUGCUCCAAAUGAAAACUUUAACCCGCCGGU ((((((.....(((((((((...))))))))).((((.......(((.((((......)))).))).......)))).............)))))) (-21.94 = -22.11 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:07 2006