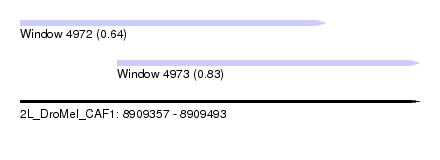
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,909,357 – 8,909,493 |
| Length | 136 |
| Max. P | 0.830394 |

| Location | 8,909,357 – 8,909,461 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
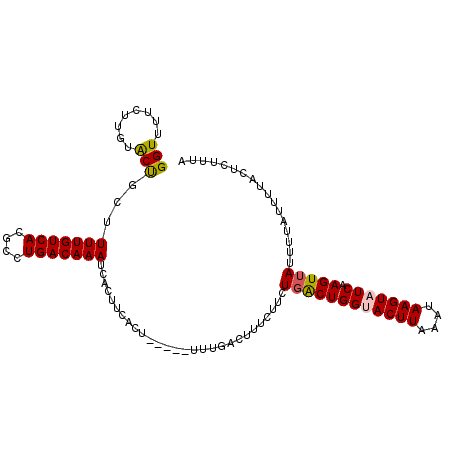
>2L_DroMel_CAF1 8909357 104 + 22407834 GGUUUUCUUGUACUGCUUUUGUCACGCCUGACAAAUCACUUCACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUA ((((....((...((..(((((((....))))))).))...))..-----...))))......((((((((((((....))))))).)))))................. ( -16.30) >DroSec_CAF1 119595 104 + 1 GGUUUUCUUGUACUGUUUUUGUCACGCCUGACAAAUCACUUAACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUA ((((.....((...((.(((((((....)))))))..))...)).-----...))))......((((((((((((....))))))).)))))................. ( -16.80) >DroSim_CAF1 118521 103 + 1 GGUUUUCUUGUACUGCAUUUGUCACGCCUGACAAAUCACUUCACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACU-UCUA ((((....((...((.((((((((....))))))))))...))..-----...))))......((((((((((((....))))))).)))))............-.... ( -18.10) >DroEre_CAF1 126956 104 + 1 GGUUUUCCUAUACCGCUUUUGUCACGCCUGACAAAUCACUUCACUUCUCUUCUGACUUUCUUCUGGCUGGGACUUAAAUAAGUAUCAAGUUAU-----UUUACUCUUUA (((........)))...(((((((....)))))))............................((((((..((((....))))..).))))).-----........... ( -12.50) >DroYak_CAF1 123502 104 + 1 GGUUUUCUUGUGCUGCCUUUGUCACGCCUGACAAAUCACUUCACU-----UCUGACUUUCUUCUCGCUGGCACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUA (((...(((((..(((((((((((....))))))).....(((..-----..))).............)))).....))))).)))....................... ( -14.10) >consensus GGUUUUCUUGUACUGCUUUUGUCACGCCUGACAAAUCACUUCACU_____UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUA (((........)))...(((((((....)))))))............................((((((((((((....))))))).)))))................. (-13.58 = -13.62 + 0.04)

| Location | 8,909,390 – 8,909,493 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -10.39 |
| Energy contribution | -11.89 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
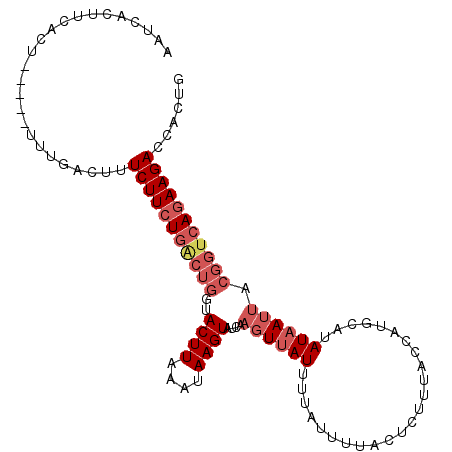
>2L_DroMel_CAF1 8909390 103 + 22407834 AAUCACUUCACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUACCAUGCAUAUAAUUACGGUCAGAAGACCAUUG ............-----........(((((((((((((((((....))))))).((((((........................))))))..))))))))))...... ( -19.96) >DroSec_CAF1 119628 103 + 1 AAUCACUUAACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUACCAUGUGUAUUAUUACGGUCAGAAGAACACUG ............-----.......((((((((((((((((((....))))))).............................(((....))))))))))))))..... ( -22.40) >DroSim_CAF1 118554 102 + 1 AAUCACUUCACU-----UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACU-UCUACCAUGUGUAUAAUUACGGUCAGAAGACCACUG ............-----........(((((((((((((((((....)))))))..................-..........(((....)))))))))))))...... ( -21.50) >DroEre_CAF1 126989 103 + 1 AAUCACUUCACUUCUCUUCUGACUUUCUUCUGGCUGGGACUUAAAUAAGUAUCAAGUUAU-----UUUACUCUUUACCGUGCAUAUAAUUACGGUCAGAAGACUACUG ..............((((((((((.......((..((((...((((((.(....).))))-----))...))))..))(((........)))))))))))))...... ( -20.60) >DroYak_CAF1 123535 103 + 1 AAUCACUUCACU-----UCUGACUUUCUUCUCGCUGGCACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUAUCGUGCAUAUAAUUACGGUCAGAAGACUACUG ..........((-----(((((((............((((...((..((((..((((......))))))))..))...))))..........)))))))))....... ( -18.75) >DroAna_CAF1 122471 83 + 1 AAUCACGCCUCU------UUGACUUUCUUCUGC------------------UCUUUUUAUUUUAUUUACCUUUUUACUGUGCAUAUAAUUACGGUCAAAAGAUUGGU- ......(((...------..((......))...------------------..................(((((.((((((........)))))).)))))...)))- ( -8.60) >consensus AAUCACUUCACU_____UUUGACUUUCUUCUGACUGGUACUUAAAUAAGUAUCAAGUUAUUUUAUUUUACUCUUUACCAUGCAUAUAAUUACGGUCAGAAGACCACUG .........................(((((((((((..((((....))))....((((((........................)))))).)))))))))))...... (-10.39 = -11.89 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:00 2006