
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,891,695 – 8,891,855 |
| Length | 160 |
| Max. P | 0.924405 |

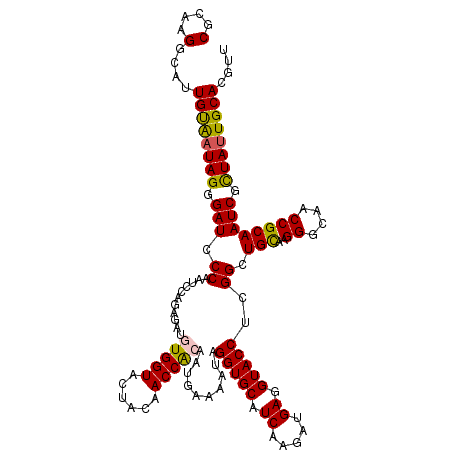
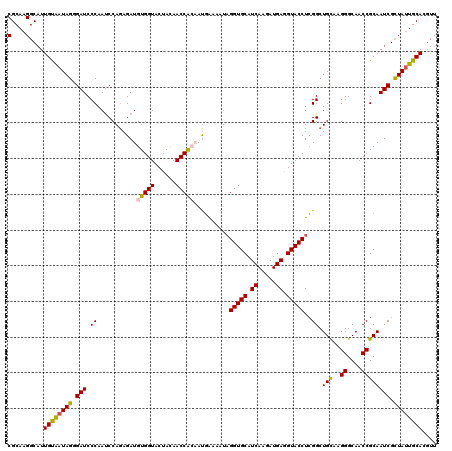
| Location | 8,891,695 – 8,891,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -36.36 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.56 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8891695 120 + 22407834 CGCAAGGCAUUGUAAUAGGGAUCCCAAUCCACAGAUGUGGUACUACAACCAACGUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGUAAGGGCAACCGCAAUCGUUAUUGCACAUU .((((((((((((....((...(((...((((....))))...((((.((...((....))((((((.((.....)).)))))).)).)))).)))...))))))).))).))))..... ( -33.00) >DroSec_CAF1 101842 120 + 1 CGCAAGGCAUUGUGAUAGGGAUCCCAAUCCAGAGAUGUGGUACUACAACCGCAAUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCGAGGGCAACCGCAAUCGCUAUUGCACGUU .((((((((((((....((....))..(((((.((((((((......)))))).........(((((.((.....)).)))))))..))).))((....))))))).))).))))..... ( -37.50) >DroSim_CAF1 100544 120 + 1 CGCAAGGCAUUGUGAUAGGGAUCCCAAUCCAGAGAUGUGGUACUACAACCACAAUGAAAAUCGGUGCAUCAAGAUGAGGUACCUCGGCUGCAAGGGCAACCGCAAUCGCUAUUGCACGUU .(((((((((((((((.((....)).)))(((.((((((((......)))))).........(((((.((.....)).)))))))..)))...((....))))))).))).))))..... ( -37.50) >DroEre_CAF1 106404 120 + 1 CGCAAGGCAUUGCAAUAGUGAUCCCAAUCCAGAGAUGUGGUACUAUAACCAAAAUCGAAAUAGGUGCAUCAAGAUGAAGUACCUGGGCUGUAAGGGCAACCGCAAUCGCUAUUGCAAGUU (....)((.((((((((((((((((......((....((((......))))...)).....((((((.((.....)).)))))))))......((....))...))))))))))))))). ( -41.10) >DroYak_CAF1 105379 120 + 1 CGCAAGGCUUUGCAUUAGAGAUCCCAACCCAGAGAUGUGGUACUAUAACCACCACGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCUUGGGCAACCGCAAUCGCUACUGCAAGUU .(((.(((.((((....(((...((...........(((((......))))).........((((((.((.....)).)))))).))...)))((....))))))..)))..)))..... ( -32.70) >consensus CGCAAGGCAUUGUAAUAGGGAUCCCAAUCCAGAGAUGUGGUACUACAACCACAAUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCAAGGGCAACCGCAAUCGCUAUUGCACGUU (....)....((((((((.(((.((...........(((((......)))))..........(((((.((.....)).)))))..)).(((..((....)))))))).)))))))).... (-31.00 = -30.56 + -0.44)

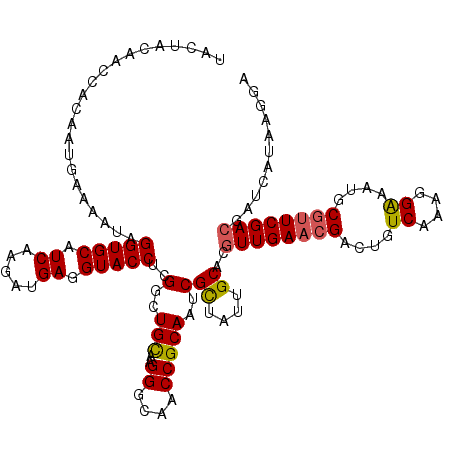
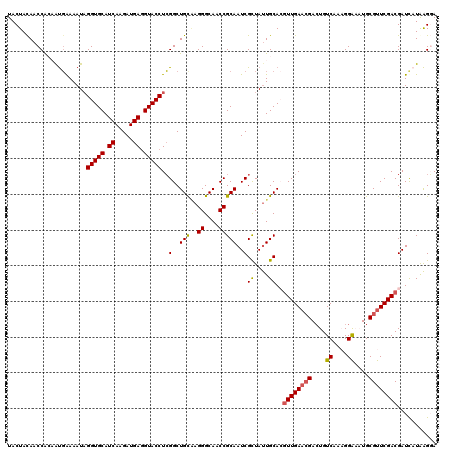
| Location | 8,891,735 – 8,891,855 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.66 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8891735 120 + 22407834 UACUACAACCAACGUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGUAAGGGCAACCGCAAUCGUUAUUGCACAUUGAACGAGUGUCAAAGGAAAUGCGUUCGACGAUCAUGAGGA ........((..(((((.....(((((.((.....)).)))))(((..(((..((....))(((((....))))))))(((((((....((....))....))))))))))))))).)). ( -32.90) >DroSec_CAF1 101882 120 + 1 UACUACAACCGCAAUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCGAGGGCAACCGCAAUCGCUAUUGCACGUUGAACGACUGUCAAAGGAAAUGCGUUCGACGAUUAUAAGGA ........((((((((.....((((((.((.....)).))))))((..(((..((....)))))..)).)))))).(((((((((....((....))....))))))))).......)). ( -40.10) >DroSim_CAF1 100584 120 + 1 UACUACAACCACAAUGAAAAUCGGUGCAUCAAGAUGAGGUACCUCGGCUGCAAGGGCAACCGCAAUCGCUAUUGCACGUUGAACGACUGUCAAAGGAAAUGCGUUCGACGAUCAUAAGGA ........((...((((.....(((((.((.....)).))))).....(((((((....))((....))..)))))(((((((((....((....))....))))))))).))))..)). ( -35.20) >DroEre_CAF1 106444 120 + 1 UACUAUAACCAAAAUCGAAAUAGGUGCAUCAAGAUGAAGUACCUGGGCUGUAAGGGCAACCGCAAUCGCUAUUGCAAGUUGAAAGACUGUCAGCGGGCCUGCGUUCGACGAUCCUAAGCA ..............(((((..((((((.((.....)).))))))(((((....((....)).....((((...(((.(((....)))))).)))))))))...)))))............ ( -33.90) >DroYak_CAF1 105419 120 + 1 UACUAUAACCACCACGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCUUGGGCAACCGCAAUCGCUACUGCAAGUUGACCGACUGUCAACGGGCCUGCGUUCGACGAUUCUAGGCA ..............((((..(((((((.(((((.((((....))))....)))))))..(((((........)))..((((((.....)))))))))))))..))))............. ( -34.10) >consensus UACUACAACCACAAUGAAAAUAGGUGCAUCAAGAUGAGGUACCUCGGCUGCAAGGGCAACCGCAAUCGCUAUUGCACGUUGAACGACUGUCAAAGGAAAUGCGUUCGACGAUCAUAAGGA ......................(((((.((.....)).)))))..(..(((..((....)))))..)((....))..((((((((....((....))....))))))))........... (-25.70 = -25.66 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:55 2006