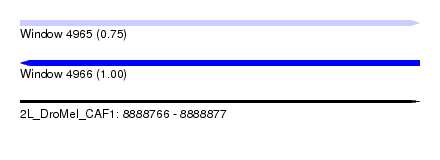
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,888,766 – 8,888,877 |
| Length | 111 |
| Max. P | 0.999629 |

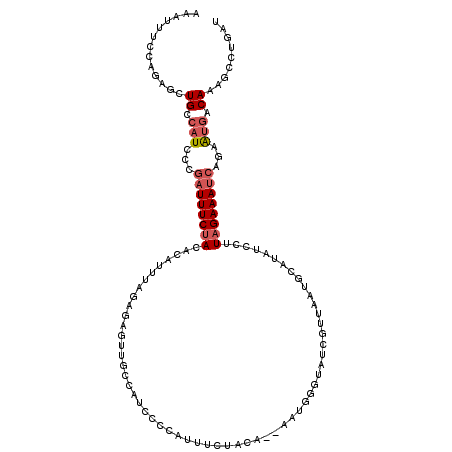
| Location | 8,888,766 – 8,888,877 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -2.35 |
| Energy contribution | -3.19 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.09 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8888766 111 + 22407834 ACAUUUCUAGAGCUGCCAUCACAAUUUCUACACAUUAAGAGAGUUGCCAUCCCGGUUUCUACA--AAUGUGUAAGAC---UACAUAUCCUUAGAAAUCAGGAUGACAAGGCCUGAU ..............(((.......(((((........))))).(((.(((((.((((((((..--.(((((((....---)))))))...)))))))).))))).))))))..... ( -29.40) >DroSec_CAF1 98837 114 + 1 ACAUUUCCAGAGCUGCCAUCCCGAUUUCUACACAUUUAGGGAGUUGCCAUCCCGACUUCUUCA--AAUAGGUAUUUUUAAUGCAUAUCAUUAGAAAUCAAAAUGACAAAGCCUGAU .......(((.(((..(((...((((((((...(((((((((((((......))))))))).)--)))..((((.....)))).......))))))))...)))....)))))).. ( -24.90) >DroSim_CAF1 97520 114 + 1 AAGUUUCCAGAGCUGCCAUCCCGAUUUCUACACAUUUAGCGAGUUGCCAUCCCCACUUCUUCA--AAUGGGUAUUUCUAAUGCAUAUCAUUAGAAAUCAAAAUGACAAAGCCUGAU .......(((.(((.......(((((((((......))).)))))).(((.((((.((....)--).)))).((((((((((.....))))))))))....)))....)))))).. ( -19.80) >DroEre_CAF1 103421 110 + 1 AAAUUUCAAGAGCUGCCAUACCGAUUUCAACACAUUACGA--GCUUUCAUCCAUUUUUAUACACACAUGCGUAUCGUUGGCG----UCCUUAGAAAACGGUGUGGCAACGCUUGAC .....(((((.(.((((((((((.((((..........(.--.....)..(((.....((((.(....).))))...)))..----......)))).)))))))))).).))))). ( -30.10) >DroYak_CAF1 102297 109 + 1 AUACAAGGAGAGCUGCCAUAUCGAUUUCUACACAUUUCGAAUGCUACCAUGCACAUUUCUACA--CAAGGGUAUCGUUAACA-----CCGUAGAAAUCAGGACUGCAACUUAUGAC .........(((.(((......(((((((((.......((((((......)))...)))....--...((((.......)).-----)))))))))))......))).)))..... ( -20.50) >consensus AAAUUUCCAGAGCUGCCAUCCCGAUUUCUACACAUUUAGAGAGUUGCCAUCCCCAUUUCUACA__AAUGGGUAUCGUUAAUGCAUAUCCUUAGAAAUCAGAAUGACAAAGCCUGAU .............((.(((...((((((((............................................................))))))))...))).))......... ( -2.35 = -3.19 + 0.84)


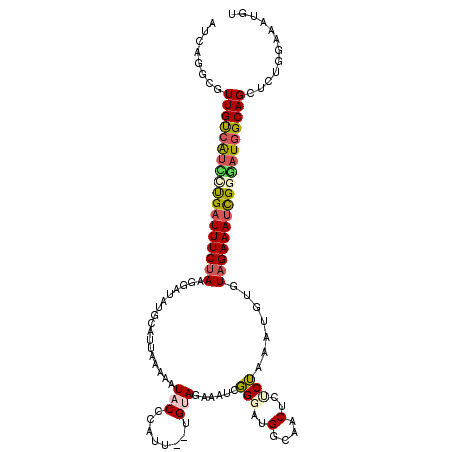
| Location | 8,888,766 – 8,888,877 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.16 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8888766 111 - 22407834 AUCAGGCCUUGUCAUCCUGAUUUCUAAGGAUAUGUA---GUCUUACACAUU--UGUAGAAACCGGGAUGGCAACUCUCUUAAUGUGUAGAAAUUGUGAUGGCAGCUCUAGAAAUGU .....((((((((((((((.((((((..(((.((((---....)))).)))--..)))))).)))))))))))...((.((((........)))).)).))).............. ( -31.30) >DroSec_CAF1 98837 114 - 1 AUCAGGCUUUGUCAUUUUGAUUUCUAAUGAUAUGCAUUAAAAAUACCUAUU--UGAAGAAGUCGGGAUGGCAACUCCCUAAAUGUGUAGAAAUCGGGAUGGCAGCUCUGGAAAUGU .(((((((..((((((((((((((((..(((...(.(((((........))--))).)..)))((((.(....)))))........))))))))))))))))))).))))...... ( -35.30) >DroSim_CAF1 97520 114 - 1 AUCAGGCUUUGUCAUUUUGAUUUCUAAUGAUAUGCAUUAGAAAUACCCAUU--UGAAGAAGUGGGGAUGGCAACUCGCUAAAUGUGUAGAAAUCGGGAUGGCAGCUCUGGAAACUU .(((((((..((((((((((((((((.......((..........((((((--(....)))))))((.(....)))))........))))))))))))))))))).))))...... ( -38.66) >DroEre_CAF1 103421 110 - 1 GUCAAGCGUUGCCACACCGUUUUCUAAGGA----CGCCAACGAUACGCAUGUGUGUAUAAAAAUGGAUGAAAGC--UCGUAAUGUGUUGAAAUCGGUAUGGCAGCUCUUGAAAUUU .(((((.(((((((.((((.((((....((----(((.....((((((....))))))....(((..(....).--.)))...))))))))).)))).))))))).)))))..... ( -37.60) >DroYak_CAF1 102297 109 - 1 GUCAUAAGUUGCAGUCCUGAUUUCUACGG-----UGUUAACGAUACCCUUG--UGUAGAAAUGUGCAUGGUAGCAUUCGAAAUGUGUAGAAAUCGAUAUGGCAGCUCUCCUUGUAU ......((((((.((..((((((((((..-----.........((((..((--..((....))..)).))))((((.....))))))))))))))..)).)))))).......... ( -30.70) >consensus AUCAGGCGUUGUCAUCCUGAUUUCUAAGGAUAUGCAUUAAAAAUACCCAUU__UGUAGAAAUCGGGAUGGCAACUCUCUAAAUGUGUAGAAAUCGGGAUGGCAGCUCUGGAAAUGU ........((((((((((((((((((.................(((........)))......(((..(....)..))).......))))))))))))))))))............ (-15.64 = -16.16 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:53 2006